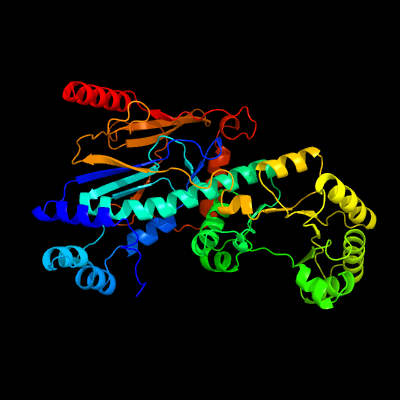
| 1 | c2f7fA_
|
|
 |
100.0 |
39 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate phosphoribosyltransferase, putative;
PDBTitle: crystal structure of enterococcus faecalis putative nicotinate2 phosphoribosyltransferase, new york structural genomics consortium
|
|
|
|
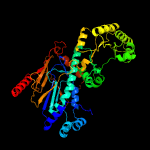
| 2 | c4yubB_
|
|
 |
100.0 |
38 |
PDB header:ligase
Chain: B: PDB Molecule:nicotinate phosphoribosyltransferase;
PDBTitle: crystal structure of human nicotinic acid phosphoribosyltransferase
|
|
|
|
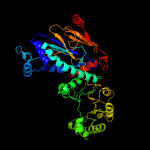
| 3 | c4hl7A_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate phosphoribosyltransferase;
PDBTitle: crystal structure of nicotinate phosphoribosyltransferase (target2 nysgr-026035) from vibrio cholerae
|
|
|
|
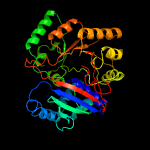
| 4 | c1yirA_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate phosphoribosyltransferase 2;
PDBTitle: crystal structure of a nicotinate phosphoribosyltransferase
|
|
|
|
| 5 | c1vlpA_
|
|
 |
100.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate phosphoribosyltransferase;
PDBTitle: crystal structure of a putative nicotinate phosphoribosyltransferase2 (yor209c, npt1) from saccharomyces cerevisiae at 1.75 a resolution
|
|
|
|
| 6 | c2im5C_
|
|
 |
100.0 |
23 |
PDB header:transferase
Chain: C: PDB Molecule:nicotinate phosphoribosyltransferase;
PDBTitle: crystal structure of nicotinate phosphoribosyltransferase from2 porphyromonas gingivalis
|
|
|
|
| 7 | c3os4A_
|
|
 |
100.0 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate phosphoribosyltransferase;
PDBTitle: the crystal structure of nicotinate phosphoribosyltransferase from2 yersinia pestis
|
|
|
|
| 8 | c1ybeA_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate phosphoribosyltransferase;
PDBTitle: crystal structure of a nicotinate phosphoribosyltransferase
|
|
|
|
| 9 | c2e5cA_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinamide phosphoribosyltransferase;
PDBTitle: crystal structure of human nmprtase complexed with 5'-phosphoribosyl-2 1'-pyrophosphate
|
|
|
|
| 10 | c2i14B_
|
|
 |
100.0 |
29 |
PDB header:transferase
Chain: B: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of nicotinate-nucleotide2 pyrophosphorylase from pyrococcus furiosus
|
|
|
|
| 11 | d2f7fa1
|
|
 |
100.0 |
38 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
|
|
|
| 12 | c1ytkA_
|
|
 |
100.0 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate phosphoribosyltransferase from thermoplasma
PDBTitle: crystal structure of a nicotinate phosphoribosyltransferase from2 thermoplasma acidophilum with nicotinate mononucleotide
|
|
|
|
| 13 | d2i14a1
|
|
 |
100.0 |
31 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
|
|
|
| 14 | d1ytda1
|
|
 |
100.0 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
|
|
|
| 15 | d1yira1
|
|
 |
100.0 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:Monomeric nicotinate phosphoribosyltransferase C-terminal domain |
|
|
|
| 16 | d1ybea1
|
|
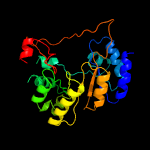 |
100.0 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:Monomeric nicotinate phosphoribosyltransferase C-terminal domain |
|
|
|
| 17 | d1vlpa2
|
|
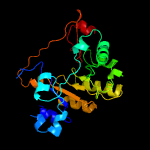 |
100.0 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:Monomeric nicotinate phosphoribosyltransferase C-terminal domain |
|
|
|
| 18 | d2f7fa2
|
|
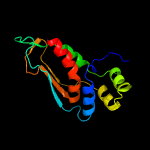 |
100.0 |
38 |
Fold:alpha/beta-Hammerhead
Superfamily:Nicotinate/Quinolinate PRTase N-terminal domain-like
Family:NadC N-terminal domain-like |
|
|
|
| 19 | d1vlpa1
|
|
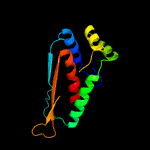 |
100.0 |
15 |
Fold:alpha/beta-Hammerhead
Superfamily:Nicotinate/Quinolinate PRTase N-terminal domain-like
Family:Monomeric nicotinate phosphoribosyltransferase N-terminal domain-like |
|
|
|
| 20 | d1yira2
|
|
 |
100.0 |
25 |
Fold:alpha/beta-Hammerhead
Superfamily:Nicotinate/Quinolinate PRTase N-terminal domain-like
Family:Monomeric nicotinate phosphoribosyltransferase N-terminal domain-like |
|
|
|
| 21 | d1ybea2 |
|
not modelled |
100.0 |
21 |
Fold:alpha/beta-Hammerhead
Superfamily:Nicotinate/Quinolinate PRTase N-terminal domain-like
Family:Monomeric nicotinate phosphoribosyltransferase N-terminal domain-like |
|
|
| 22 | c2b7pA_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:probable nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of quinolinic acid phosphoribosyltransferase from2 helicobacter pylori with phthalic acid
|
|
|
| 23 | c1x1oC_ |
|
not modelled |
100.0 |
22 |
PDB header:transferase
Chain: C: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of project id tt0268 from thermus thermophilus hb8
|
|
|
| 24 | c1qpoA_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:quinolinate acid phosphoribosyl transferase;
PDBTitle: quinolinate phosphoribosyl transferase (qaprtase) apo-enzyme from2 mycobacterium tuberculosis
|
|
|
| 25 | c3l0gD_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: D: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of nicotinate-nucleotide pyrophosphorylase from2 ehrlichia chaffeensis at 2.05a resolution
|
|
|
| 26 | c2jbmA_ |
|
not modelled |
100.0 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: qprtase structure from human
|
|
|
| 27 | c3pajA_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide pyrophosphorylase, carboxylating;
PDBTitle: 2.00 angstrom resolution crystal structure of a quinolinate2 phosphoribosyltransferase from vibrio cholerae o1 biovar eltor str.3 n16961
|
|
|
| 28 | c5huoH_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: H: PDB Molecule:nicotinate-nucleotide diphosphorylase (carboxylating);
PDBTitle: crystal structure of nadc deletion mutant in c2221 space group
|
|
|
| 29 | c1o4uA_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:type ii quinolic acid phosphoribosyltransferase;
PDBTitle: crystal structure of a nicotinate nucleotide pyrophosphorylase2 (tm1645) from thermotoga maritima at 2.50 a resolution
|
|
|
| 30 | c1qapA_ |
|
not modelled |
100.0 |
21 |
PDB header:glycosyltransferase
Chain: A: PDB Molecule:quinolinic acid phosphoribosyltransferase;
PDBTitle: quinolinic acid phosphoribosyltransferase with bound2 quinolinic acid
|
|
|
| 31 | c3tqvA_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: structure of the nicotinate-nucleotide pyrophosphorylase from2 francisella tularensis.
|
|
|
| 32 | c3gnnA_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of nicotinate-nucleotide pyrophosphorylase from2 burkholderi pseudomallei
|
|
|
| 33 | c3c2vA_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of the quinolinate phosphoribosyl transferase (bna6)2 from saccharomyces cerevisiae complexed with prpp and the inhibitor3 phthalate
|
|
|
| 34 | d2i14a2 |
|
not modelled |
100.0 |
22 |
Fold:alpha/beta-Hammerhead
Superfamily:Nicotinate/Quinolinate PRTase N-terminal domain-like
Family:NadC N-terminal domain-like |
|
|
| 35 | d1ytda2 |
|
not modelled |
100.0 |
27 |
Fold:alpha/beta-Hammerhead
Superfamily:Nicotinate/Quinolinate PRTase N-terminal domain-like
Family:NadC N-terminal domain-like |
|
|
| 36 | d1qapa1 |
|
not modelled |
99.8 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
|
|
| 37 | d1qpoa1 |
|
not modelled |
99.8 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
|
|
| 38 | d1o4ua1 |
|
not modelled |
99.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
|
|
| 39 | d1qpoa2 |
|
not modelled |
97.7 |
22 |
Fold:alpha/beta-Hammerhead
Superfamily:Nicotinate/Quinolinate PRTase N-terminal domain-like
Family:NadC N-terminal domain-like |
|
|
| 40 | d1qapa2 |
|
not modelled |
97.4 |
26 |
Fold:alpha/beta-Hammerhead
Superfamily:Nicotinate/Quinolinate PRTase N-terminal domain-like
Family:NadC N-terminal domain-like |
|
|
| 41 | d1o4ua2 |
|
not modelled |
96.9 |
16 |
Fold:alpha/beta-Hammerhead
Superfamily:Nicotinate/Quinolinate PRTase N-terminal domain-like
Family:NadC N-terminal domain-like |
|
|
| 42 | c6oviA_ |
|
not modelled |
93.2 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:keto-deoxy-phosphogluconate aldolase;
PDBTitle: crystal structure of kdpg aldolase from legionella pneumophila with2 pyruvate captured at low ph as a covalent carbinolamine intermediate
|
|
|
| 43 | c3cu2A_ |
|
not modelled |
90.8 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:ribulose-5-phosphate 3-epimerase;
PDBTitle: crystal structure of ribulose-5-phosphate 3-epimerase (yp_718263.1)2 from haemophilus somnus 129pt at 1.91 a resolution
|
|
|
| 44 | d7reqa2 |
|
not modelled |
90.6 |
20 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 45 | c1e1cA_ |
|
not modelled |
89.6 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:methylmalonyl-coa mutase alpha chain;
PDBTitle: methylmalonyl-coa mutase h244a mutant
|
|
|
| 46 | d2flia1 |
|
not modelled |
88.1 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 47 | c5umfB_ |
|
not modelled |
88.0 |
22 |
PDB header:isomerase
Chain: B: PDB Molecule:ribulose-phosphate 3-epimerase;
PDBTitle: crystal structure of a ribulose-phosphate 3-epimerase from neisseria2 gonorrhoeae with bound phosphate
|
|
|
| 48 | c3tsmB_ |
|
not modelled |
87.6 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of indole-3-glycerol phosphate synthase from2 brucella melitensis
|
|
|
| 49 | d1xi3a_ |
|
not modelled |
87.6 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Thiamin phosphate synthase
Family:Thiamin phosphate synthase |
|
|
| 50 | d1tqxa_ |
|
not modelled |
87.3 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 51 | c4bk9B_ |
|
not modelled |
86.5 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxo
PDBTitle: crystal structure of 2-keto-3-deoxy-6-phospho-gluconate aldolase from2 zymomonas mobilis atcc 29191
|
|
|
| 52 | c4qccA_ |
|
not modelled |
86.2 |
28 |
PDB header:structural protein, lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxy-6-phosphogalactonate aldolase, peptidyl-
PDBTitle: structure of a cube-shaped, highly porous protein cage designed by2 fusing symmetric oligomeric domains
|
|
|
| 53 | d1rpxa_ |
|
not modelled |
85.5 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 54 | d1ea0a2 |
|
not modelled |
84.3 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 55 | c5aheA_ |
|
not modelled |
83.8 |
33 |
PDB header:isomerase
Chain: A: PDB Molecule:
PDBTitle: crystal structure of salmonella enterica hisa
|
|
|
| 56 | c3inpA_ |
|
not modelled |
83.8 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:d-ribulose-phosphate 3-epimerase;
PDBTitle: 2.05 angstrom resolution crystal structure of d-ribulose-phosphate 3-2 epimerase from francisella tularensis.
|
|
|
| 57 | d1hg3a_ |
|
not modelled |
83.1 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 58 | c3o63B_ |
|
not modelled |
82.5 |
26 |
PDB header:transferase
Chain: B: PDB Molecule:probable thiamine-phosphate pyrophosphorylase;
PDBTitle: crystal structure of thiamin phosphate synthase from mycobacterium2 tuberculosis
|
|
|
| 59 | d1vhca_ |
|
not modelled |
82.4 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 60 | c2v82A_ |
|
not modelled |
81.9 |
26 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxy-6-phosphogalactonate aldolase;
PDBTitle: kdpgal complexed to kdpgal
|
|
|
| 61 | c4e38A_ |
|
not modelled |
81.7 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:keto-hydroxyglutarate-aldolase/keto-deoxy-phosphogluconate
PDBTitle: crystal structure of probable keto-hydroxyglutarate-aldolase from2 vibrionales bacterium swat-3 (target efi-502156)
|
|
|
| 62 | d2tpsa_ |
|
not modelled |
81.5 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Thiamin phosphate synthase
Family:Thiamin phosphate synthase |
|
|
| 63 | d1a53a_ |
|
not modelled |
81.5 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 64 | c3labA_ |
|
not modelled |
80.8 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative kdpg (2-keto-3-deoxy-6-phosphogluconate) aldolase;
PDBTitle: crystal structure of a putative kdpg (2-keto-3-deoxy-6-2 phosphogluconate) aldolase from oleispira antarctica
|
|
|
| 65 | d1ofda2 |
|
not modelled |
80.7 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 66 | c1yadD_ |
|
not modelled |
80.3 |
19 |
PDB header:transcription
Chain: D: PDB Molecule:regulatory protein teni;
PDBTitle: structure of teni from bacillus subtilis
|
|
|
| 67 | d1wbha1 |
|
not modelled |
80.2 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 68 | c2h6rG_ |
|
not modelled |
79.8 |
18 |
PDB header:isomerase
Chain: G: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase (tim) from2 methanocaldococcus jannaschii
|
|
|
| 69 | c2c3zA_ |
|
not modelled |
79.0 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of a truncated variant of indole-3-2 glycerol phosphate synthase from sulfolobus solfataricus
|
|
|
| 70 | c3qjaA_ |
|
not modelled |
75.6 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of the mycobacterium tuberculosis indole-3-glycerol2 phosphate synthase (trpc) in apo form
|
|
|
| 71 | d1wa3a1 |
|
not modelled |
75.4 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 72 | c4utwB_ |
|
not modelled |
73.9 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:putative n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structural characterisation of nane, mannac6p c2 epimerase,2 from clostridium perfingens
|
|
|
| 73 | c3qc3B_ |
|
not modelled |
73.6 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:d-ribulose-5-phosphate-3-epimerase;
PDBTitle: crystal structure of a d-ribulose-5-phosphate-3-epimerase (np_954699)2 from homo sapiens at 2.20 a resolution
|
|
|
| 74 | c4nu7C_ |
|
not modelled |
72.6 |
21 |
PDB header:isomerase
Chain: C: PDB Molecule:ribulose-phosphate 3-epimerase;
PDBTitle: 2.05 angstrom crystal structure of ribulose-phosphate 3-epimerase from2 toxoplasma gondii.
|
|
|
| 75 | c3ceuA_ |
|
not modelled |
72.2 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:thiamine phosphate pyrophosphorylase;
PDBTitle: crystal structure of thiamine phosphate pyrophosphorylase (bt_0647)2 from bacteroides thetaiotaomicron. northeast structural genomics3 consortium target btr268
|
|
|
| 76 | d1tqja_ |
|
not modelled |
71.9 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 77 | c5n2pA_ |
|
not modelled |
70.9 |
28 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: sulfolobus solfataricus tryptophan synthase a
|
|
|
| 78 | c6bmaA_ |
|
not modelled |
70.8 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: the crystal structure of indole-3-glycerol phosphate synthase from2 campylobacter jejuni subsp. jejuni nctc 11168
|
|
|
| 79 | d1mxsa_ |
|
not modelled |
69.8 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 80 | c3ct7E_ |
|
not modelled |
67.2 |
21 |
PDB header:isomerase
Chain: E: PDB Molecule:d-allulose-6-phosphate 3-epimerase;
PDBTitle: crystal structure of d-allulose 6-phosphate 3-epimerase2 from escherichia coli k-12
|
|
|
| 81 | d1rd5a_ |
|
not modelled |
67.1 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 82 | d1ccwa_ |
|
not modelled |
67.1 |
20 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 83 | d1vzwa1 |
|
not modelled |
66.9 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
| 84 | d1h5ya_ |
|
not modelled |
66.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
| 85 | d1w0ma_ |
|
not modelled |
66.4 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 86 | d1thfd_ |
|
not modelled |
66.3 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
| 87 | c3igsB_ |
|
not modelled |
65.9 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase 2;
PDBTitle: structure of the salmonella enterica n-acetylmannosamine-6-phosphate2 2-epimerase
|
|
|
| 88 | c4n6eA_ |
|
not modelled |
64.8 |
15 |
PDB header:lyase/biosynthetic protein
Chain: A: PDB Molecule:putative thiosugar synthase;
PDBTitle: crystal structure of amycolatopsis orientalis bexx/cyso complex
|
|
|
| 89 | d1jgta1 |
|
not modelled |
64.7 |
31 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
|
|
| 90 | c5cssA_ |
|
not modelled |
64.6 |
10 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from thermoplasma2 acidophilum with glycerol 3-phosphate
|
|
|
| 91 | c3gr7A_ |
|
not modelled |
63.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase;
PDBTitle: structure of oye from geobacillus kaustophilus, hexagonal crystal form
|
|
|
| 92 | d1h1ya_ |
|
not modelled |
63.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 93 | c5zknA_ |
|
not modelled |
62.8 |
10 |
PDB header:isomerase
Chain: A: PDB Molecule:putative n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate 2-epimerase from2 fusobacterium nucleatum
|
|
|
| 94 | c4x2rA_ |
|
not modelled |
61.9 |
31 |
PDB header:isomerase
Chain: A: PDB Molecule:1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)
PDBTitle: crystal structure of pria from actinomyces urogenitalis
|
|
|
| 95 | c3f4wA_ |
|
not modelled |
60.6 |
18 |
PDB header:synthase, lyase
Chain: A: PDB Molecule:putative hexulose 6 phosphate synthase;
PDBTitle: the 1.65a crystal structure of 3-hexulose-6-phosphate2 synthase from salmonella typhimurium
|
|
|
| 96 | d1qopa_ |
|
not modelled |
59.9 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 97 | c4g1kB_ |
|
not modelled |
58.8 |
22 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from burkholderia2 thailandensis
|
|
|
| 98 | c5zjnB_ |
|
not modelled |
57.2 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:putative n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate-2-epimerase from vibrio2 cholerae with n-acetylmannosamine-6-phosphate
|
|
|
| 99 | c4axkB_ |
|
not modelled |
57.2 |
27 |
PDB header:isomerase
Chain: B: PDB Molecule:1-(5-phosphoribosyl)-5-((5'-phosphoribosylamino)
PDBTitle: crystal structure of subhisa from the thermophile corynebacterium2 efficiens
|
|
|
| 100 | c3absD_ |
|
not modelled |
56.2 |
26 |
PDB header:lyase
Chain: D: PDB Molecule:ethanolamine ammonia-lyase light chain;
PDBTitle: crystal structure of ethanolamine ammonia-lyase from escherichia coli2 complexed with adeninylpentylcobalamin and ethanolamine
|
|
|
| 101 | d1xcfa_ |
|
not modelled |
55.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 102 | d1yxya1 |
|
not modelled |
55.4 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:NanE-like |
|
|
| 103 | d1ka9f_ |
|
not modelled |
55.3 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
| 104 | c5b69A_ |
|
not modelled |
54.6 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:geranylgeranylglyceryl phosphate synthase;
PDBTitle: crystal structure of geranylgeranylglyceryl phosphate synthase2 complexed with an g-1-p from thermoplasma acidophilum
|
|
|
| 105 | c2bdqA_ |
|
not modelled |
54.2 |
17 |
PDB header:metal transport
Chain: A: PDB Molecule:copper homeostasis protein cutc;
PDBTitle: crystal structure of the putative copper homeostasis protein cutc from2 streptococcus agalactiae, northeast strucural genomics target sar15.
|
|
|
| 106 | d1geqa_ |
|
not modelled |
54.2 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 107 | c4ml9A_ |
|
not modelled |
54.1 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized tim barrel protein with the2 conserved phosphate binding site fromsebaldella termitidis
|
|
|
| 108 | c3tdmD_ |
|
not modelled |
53.8 |
25 |
PDB header:de novo protein
Chain: D: PDB Molecule:computationally designed two-fold symmetric tim-barrel
PDBTitle: computationally designed tim-barrel protein, halfflr
|
|
|
| 109 | c1znnF_ |
|
not modelled |
53.7 |
20 |
PDB header:biosynthetic protein
Chain: F: PDB Molecule:plp synthase;
PDBTitle: structure of the synthase subunit of plp synthase
|
|
|
| 110 | d1znna1 |
|
not modelled |
53.7 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:PdxS-like |
|
|
| 111 | c6nkeA_ |
|
not modelled |
53.7 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:geranylgeranylglyceryl phosphate synthase;
PDBTitle: wild-type gggps from thermoplasma volcanium
|
|
|
| 112 | c2yw3E_ |
|
not modelled |
53.3 |
20 |
PDB header:lyase
Chain: E: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase/2-deydro-3-
PDBTitle: crystal structure analysis of the 4-hydroxy-2-oxoglutarate aldolase/2-2 deydro-3-deoxyphosphogluconate aldolase from tthb1
|
|
|
| 113 | c3bicA_ |
|
not modelled |
51.5 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:methylmalonyl-coa mutase, mitochondrial precursor;
PDBTitle: crystal structure of human methylmalonyl-coa mutase
|
|
|
| 114 | c6bkaA_ |
|
not modelled |
51.2 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nitronate monooxygenase;
PDBTitle: crystal structure of nitronate monooxygenase from cyberlindnera2 saturnus
|
|
|
| 115 | c3tdnB_ |
|
not modelled |
50.7 |
25 |
PDB header:de novo protein
Chain: B: PDB Molecule:flr symmetric alpha-beta tim barrel;
PDBTitle: computationally designed two-fold symmetric tim-barrel protein, flr
|
|
|
| 116 | d1gwja_ |
|
not modelled |
50.1 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 117 | d1vc4a_ |
|
not modelled |
49.1 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 118 | c2e18B_ |
|
not modelled |
48.4 |
24 |
PDB header:ligase
Chain: B: PDB Molecule:nh(3)-dependent nad(+) synthetase;
PDBTitle: crystal structure of project ph0182 from pyrococcus horikoshii ot3
|
|
|
| 119 | c5ocsB_ |
|
not modelled |
46.5 |
14 |
PDB header:flavoprotein
Chain: B: PDB Molecule:putative nadh-depentdent flavin oxidoreductase;
PDBTitle: ene-reductase (er/oye) from ralstonia (cupriavidus) metallidurans
|
|
|
| 120 | c2jgqB_ |
|
not modelled |
45.5 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: kinetics and structural properties of triosephosphate2 isomerase from helicobacter pylori
|
|
|