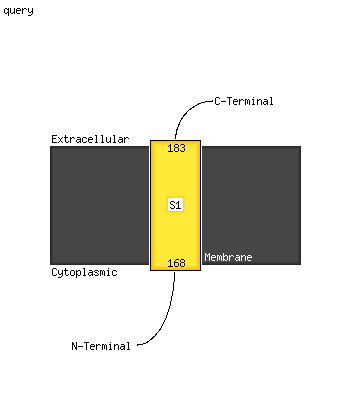
| 1 | d1t70a_
|
|
 |
100.0 |
17 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:DR1281-like |
|
|
|
| 2 | d1t71a_
|
|
 |
100.0 |
18 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:DR1281-like |
|
|
|
| 3 | c4b2oB_
|
|
 |
100.0 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:ymdb phosphodiesterase;
PDBTitle: crystal structure of bacillus subtilis ymdb, a global2 regulator of late adaptive responses.
|
|
|
|
| 4 | d2z06a1
|
|
 |
99.9 |
26 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:TTHA0625-like |
|
|
|
| 5 | d1usha2
|
|
 |
99.8 |
15 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:5'-nucleotidase (syn. UDP-sugar hydrolase), N-terminal domain |
|
|
|
| 6 | d2z1aa2
|
|
 |
99.8 |
18 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:5'-nucleotidase (syn. UDP-sugar hydrolase), N-terminal domain |
|
|
|
| 7 | c3jyfB_
|
|
 |
99.8 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:2',3'-cyclic nucleotide 2'-phosphodiesterase/3'-
PDBTitle: the crystal structure of a 2,3-cyclic nucleotide 2-2 phosphodiesterase/3-nucleotidase bifunctional periplasmic precursor3 protein from klebsiella pneumoniae subsp. pneumoniae mgh 78578
|
|
|
|
| 8 | c3ivdA_
|
|
 |
99.8 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:nucleotidase;
PDBTitle: putative 5'-nucleotidase (c4898) from escherichia coli in complex with2 uridine
|
|
|
|
| 9 | c1oidA_
|
|
 |
99.7 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein usha;
PDBTitle: 5'-nucleotidase (e. coli) with an engineered disulfide2 bridge (s228c, p513c)
|
|
|
|
| 10 | c3gveB_
|
|
 |
99.7 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:yfkn protein;
PDBTitle: crystal structure of calcineurin-like phosphoesterase yfkn from2 bacillus subtilis
|
|
|
|
| 11 | c3qfkA_
|
|
 |
99.7 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: 2.05 angstrom crystal structure of putative 5'-nucleotidase from2 staphylococcus aureus in complex with alpha-ketoglutarate
|
|
|
|
| 12 | c2z1aA_
|
|
 |
99.7 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:5'-nucleotidase;
PDBTitle: crystal structure of 5'-nucleotidase precursor from thermus2 thermophilus hb8
|
|
|
|
| 13 | c2wdfA_
|
|
 |
99.6 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:sulfur oxidation protein soxb;
PDBTitle: termus thermophilus sulfate thiohydrolase soxb
|
|
|
|
| 14 | c3zu0A_
|
|
 |
99.6 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:nad nucleotidase;
PDBTitle: structure of haemophilus influenzae nad nucleotidase (nadn)
|
|
|
|
| 15 | c4h1sB_
|
|
 |
99.6 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:5'-nucleotidase;
PDBTitle: crystal structure of a truncated soluble form of human cd73 with ecto-2 5'-nucleotidase activity
|
|
|
|
| 16 | c5h7wB_
|
|
 |
99.5 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:venom 5'-nucleotidase;
PDBTitle: crystal structure of 5'-nucleotidase from venom of naja atra
|
|
|
|
| 17 | c3c9fB_
|
|
 |
99.2 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:5'-nucleotidase;
PDBTitle: crystal structure of 5'-nucleotidase from candida albicans sc5314
|
|
|
|
| 18 | d3c9fa2
|
|
 |
99.2 |
12 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:5'-nucleotidase (syn. UDP-sugar hydrolase), N-terminal domain |
|
|
|
| 19 | c2q8uA_
|
|
 |
97.6 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:exonuclease, putative;
PDBTitle: crystal structure of mre11 from thermotoga maritima msb8 (tm1635) at2 2.20 a resolution
|
|
|
|
| 20 | c3qg5D_
|
|
 |
97.4 |
16 |
PDB header:hydrolase
Chain: D: PDB Molecule:mre11;
PDBTitle: the mre11:rad50 complex forms an atp dependent molecular clamp in dna2 double-strand break repair
|
|
|
|
| 21 | c4ltyD_ |
|
not modelled |
97.4 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:exonuclease subunit sbcd;
PDBTitle: crystal structure of e.coli sbcd at 1.8 a resolution
|
|
|
| 22 | c4ykeA_ |
|
not modelled |
97.2 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:mre11;
PDBTitle: crystal structure of eukaryotic mre11 catalytic domain from chaetomium2 thermophilum
|
|
|
| 23 | d1ii7a_ |
|
not modelled |
96.9 |
14 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:DNA double-strand break repair nuclease |
|
|
| 24 | c3av0A_ |
|
not modelled |
96.8 |
16 |
PDB header:recombination
Chain: A: PDB Molecule:dna double-strand break repair protein mre11;
PDBTitle: crystal structure of mre11-rad50 bound to atp s
|
|
|
| 25 | c3t1iC_ |
|
not modelled |
96.5 |
20 |
PDB header:hydrolase
Chain: C: PDB Molecule:double-strand break repair protein mre11a;
PDBTitle: crystal structure of human mre11: understanding tumorigenic mutations
|
|
|
| 26 | c4fbkB_ |
|
not modelled |
96.5 |
20 |
PDB header:hydrolase, protein binding
Chain: B: PDB Molecule:dna repair and telomere maintenance protein nbs1,dna repair
PDBTitle: crystal structure of a covalently fused nbs1-mre11 complex with one2 manganese ion per active site
|
|
|
| 27 | c3auzA_ |
|
not modelled |
96.1 |
16 |
PDB header:recombination
Chain: A: PDB Molecule:dna double-strand break repair protein mre11;
PDBTitle: crystal structure of mre11 with manganese
|
|
|
| 28 | c2ejcA_ |
|
not modelled |
95.7 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:pantoate--beta-alanine ligase;
PDBTitle: crystal structure of pantoate--beta-alanine ligase (panc)2 from thermotoga maritima
|
|
|
| 29 | c2xmoB_ |
|
not modelled |
95.6 |
10 |
PDB header:hydrolase
Chain: B: PDB Molecule:lmo2642 protein;
PDBTitle: the crystal structure of lmo2642
|
|
|
| 30 | c4fcxB_ |
|
not modelled |
95.5 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:dna repair protein rad32;
PDBTitle: s.pombe mre11 apoenzym
|
|
|
| 31 | c3tghA_ |
|
not modelled |
95.0 |
21 |
PDB header:cell invasion
Chain: A: PDB Molecule:glideosome-associated protein 50;
PDBTitle: gap50 the anchor in the inner membrane complex of plasmodium
|
|
|
| 32 | c3qfnA_ |
|
not modelled |
94.8 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of streptococcal asymmetric ap4a hydrolase and2 phosphodiesterase spr1479/saph in complex with inorganic phosphate
|
|
|
| 33 | d1v8fa_ |
|
not modelled |
94.5 |
23 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Pantothenate synthetase (Pantoate-beta-alanine ligase, PanC) |
|
|
| 34 | c3n8hA_ |
|
not modelled |
94.4 |
12 |
PDB header:ligase
Chain: A: PDB Molecule:pantothenate synthetase;
PDBTitle: crystal structure of pantoate-beta-alanine ligase from francisella2 tularensis
|
|
|
| 35 | c3l23A_ |
|
not modelled |
94.3 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:sugar phosphate isomerase/epimerase;
PDBTitle: crystal structure of sugar phosphate isomerase/epimerase2 (yp_001303399.1) from parabacteroides distasonis atcc 8503 at 1.70 a3 resolution
|
|
|
| 36 | d1ihoa_ |
|
not modelled |
94.1 |
18 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Pantothenate synthetase (Pantoate-beta-alanine ligase, PanC) |
|
|
| 37 | c1xzwB_ |
|
not modelled |
94.1 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:purple acid phosphatase;
PDBTitle: sweet potato purple acid phosphatase/phosphate complex
|
|
|
| 38 | c3zk4A_ |
|
not modelled |
93.9 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:diphosphonucleotide phosphatase 1;
PDBTitle: structure of purple acid phosphatase ppd1 isolated from2 yellow lupin (lupinus luteus) seeds
|
|
|
| 39 | d2qfra2 |
|
not modelled |
93.9 |
16 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Purple acid phosphatase-like |
|
|
| 40 | c2hk1D_ |
|
not modelled |
93.8 |
15 |
PDB header:isomerase
Chain: D: PDB Molecule:d-psicose 3-epimerase;
PDBTitle: crystal structure of d-psicose 3-epimerase (dpease) in the presence of2 d-fructose
|
|
|
| 41 | c1kbpB_ |
|
not modelled |
93.6 |
13 |
PDB header:hydrolase (phosphoric monoester)
Chain: B: PDB Molecule:purple acid phosphatase;
PDBTitle: kidney bean purple acid phosphatase
|
|
|
| 42 | c3ib7A_ |
|
not modelled |
93.4 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:icc protein;
PDBTitle: crystal structure of full length rv0805
|
|
|
| 43 | c3ag5A_ |
|
not modelled |
93.0 |
23 |
PDB header:ligase
Chain: A: PDB Molecule:pantothenate synthetase;
PDBTitle: crystal structure of pantothenate synthetase from staphylococcus2 aureus
|
|
|
| 44 | c3uk2B_ |
|
not modelled |
92.5 |
9 |
PDB header:ligase
Chain: B: PDB Molecule:pantothenate synthetase;
PDBTitle: the structure of pantothenate synthetase from burkholderia2 thailandensis
|
|
|
| 45 | d1xzwa2 |
|
not modelled |
91.8 |
15 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Purple acid phosphatase-like |
|
|
| 46 | c2ou4C_ |
|
not modelled |
91.2 |
9 |
PDB header:isomerase
Chain: C: PDB Molecule:d-tagatose 3-epimerase;
PDBTitle: crystal structure of d-tagatose 3-epimerase from2 pseudomonas cichorii
|
|
|
| 47 | d1uf3a_ |
|
not modelled |
91.0 |
13 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:TT1561-like |
|
|
| 48 | d1rpxa_ |
|
not modelled |
90.4 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 49 | c3innB_ |
|
not modelled |
90.0 |
16 |
PDB header:ligase
Chain: B: PDB Molecule:pantothenate synthetase;
PDBTitle: crystal structure of pantoate-beta-alanine-ligase in complex2 with atp at low occupancy at 2.1 a resolution
|
|
|
| 50 | d2a84a1 |
|
not modelled |
90.0 |
26 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Pantothenate synthetase (Pantoate-beta-alanine ligase, PanC) |
|
|
| 51 | c3mxtA_ |
|
not modelled |
89.7 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:pantothenate synthetase;
PDBTitle: crystal structure of pantoate-beta-alanine ligase from campylobacter2 jejuni
|
|
|
| 52 | d2yvta1 |
|
not modelled |
89.3 |
9 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:TT1561-like |
|
|
| 53 | d1gqna_ |
|
not modelled |
89.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 54 | c5y0tD_ |
|
not modelled |
88.2 |
20 |
PDB header:ligase
Chain: D: PDB Molecule:thermotoga maritima tmcal;
PDBTitle: crystal structure of thermotoga maritima tmcal bound with alpha-thio2 atp(form ii)
|
|
|
| 55 | c5y0nB_ |
|
not modelled |
87.9 |
17 |
PDB header:ligase
Chain: B: PDB Molecule:upf0348 protein b4417_3650;
PDBTitle: crystal structure of bacillus subtilis tmcal bound with atp (semet2 derivative)
|
|
|
| 56 | c3rmjB_ |
|
not modelled |
87.7 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of truncated alpha-isopropylmalate synthase from2 neisseria meningitidis
|
|
|
| 57 | c3vniC_ |
|
not modelled |
85.4 |
13 |
PDB header:isomerase
Chain: C: PDB Molecule:xylose isomerase domain protein tim barrel;
PDBTitle: crystal structures of d-psicose 3-epimerase from clostridium2 cellulolyticum h10 and its complex with ketohexose sugars
|
|
|
| 58 | c2yr1B_ |
|
not modelled |
85.3 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of 3-dehydroquinate dehydratase from geobacillus2 kaustophilus hta426
|
|
|
| 59 | c3guzB_ |
|
not modelled |
84.6 |
19 |
PDB header:ligase
Chain: B: PDB Molecule:pantothenate synthetase;
PDBTitle: structural and substrate-binding studies of pantothenate2 synthenate (ps)provide insights into homotropic inhibition3 by pantoate in ps's
|
|
|
| 60 | c5kwvA_ |
|
not modelled |
84.6 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:pantothenate synthetase;
PDBTitle: crystal structure of a pantoate-beta-alanine ligase from neisseria2 gonorrhoeae with bound amppnp
|
|
|
| 61 | c4utwB_ |
|
not modelled |
84.0 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:putative n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structural characterisation of nane, mannac6p c2 epimerase,2 from clostridium perfingens
|
|
|
| 62 | d1sfla_ |
|
not modelled |
83.9 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 63 | c5xceB_ |
|
not modelled |
83.7 |
17 |
PDB header:transport protein
Chain: B: PDB Molecule:vacuolar protein sorting-associated protein 29;
PDBTitle: crystal structure of wild type vps29 from entamoeba histolytica
|
|
|
| 64 | c3l2iB_ |
|
not modelled |
83.6 |
23 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.85 angstrom crystal structure of the 3-dehydroquinate dehydratase2 (arod) from salmonella typhimurium lt2.
|
|
|
| 65 | c3dx5A_ |
|
not modelled |
83.6 |
6 |
PDB header:lyase
Chain: A: PDB Molecule:uncharacterized protein asbf;
PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
|
|
|
| 66 | c4madA_ |
|
not modelled |
83.2 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-galactosidase;
PDBTitle: crystal structure of beta-galactosidase c (bgac) from bacillus2 circulans atcc 31382
|
|
|
| 67 | c4ovxA_ |
|
not modelled |
82.3 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:xylose isomerase domain protein tim barrel;
PDBTitle: crystal structure of xylose isomerase domain protein from planctomyces2 limnophilus dsm 3776
|
|
|
| 68 | c3wqoB_ |
|
not modelled |
82.1 |
5 |
PDB header:unknown function
Chain: B: PDB Molecule:uncharacterized protein mj1311;
PDBTitle: crystal structure of d-tagatose 3-epimerase-like protein
|
|
|
| 69 | d1z2wa1 |
|
not modelled |
82.0 |
15 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:YfcE-like |
|
|
| 70 | c3ewbX_ |
|
not modelled |
81.7 |
9 |
PDB header:transferase
Chain: X: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of n-terminal domain of putative 2-isopropylmalate2 synthase from listeria monocytogenes
|
|
|
| 71 | c5tenH_ |
|
not modelled |
81.2 |
11 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:4-hydroxy-tetrahydrodipicolinate reductase;
PDBTitle: structure of 4-hydroxy-tetrahydrodipicolinate reductase from vibrio2 vulnificus with 2,5 furan dicarboxylic and nadh with intact3 polyhistidine tag
|
|
|
| 72 | c4ph6A_ |
|
not modelled |
80.7 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: structure of 3-dehydroquinate dehydratase from enterococcus faecalis
|
|
|
| 73 | c5n2pA_ |
|
not modelled |
80.4 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: sulfolobus solfataricus tryptophan synthase a
|
|
|
| 74 | c3js3C_ |
|
not modelled |
79.6 |
16 |
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of type i 3-dehydroquinate dehydratase (arod) from2 clostridium difficile with covalent reaction intermediate
|
|
|
| 75 | c3gmiA_ |
|
not modelled |
79.1 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0348 protein mj0951;
PDBTitle: crystal structure of a protein of unknown function from2 methanocaldococcus jannaschii
|
|
|
| 76 | d3ck2a1 |
|
not modelled |
77.9 |
11 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:YfcE-like |
|
|
| 77 | c5hmqE_ |
|
not modelled |
77.6 |
16 |
PDB header:lyase
Chain: E: PDB Molecule:4-hydroxyphenylpyruvate dioxygenase;
PDBTitle: xylose isomerase-like tim barrel/4-hydroxyphenylpyruvate dioxygenase2 fusion protein
|
|
|
| 78 | c2nydB_ |
|
not modelled |
76.5 |
11 |
PDB header:unknown function
Chain: B: PDB Molecule:upf0135 protein sa1388;
PDBTitle: crystal structure of staphylococcus aureus hypothetical protein sa1388
|
|
|
| 79 | c2gx8B_ |
|
not modelled |
76.4 |
15 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:nif3-related protein;
PDBTitle: the crystal structure of bacillus cereus protein related to nif3
|
|
|
| 80 | d2gx8a1 |
|
not modelled |
76.2 |
15 |
Fold:NIF3 (NGG1p interacting factor 3)-like
Superfamily:NIF3 (NGG1p interacting factor 3)-like
Family:NIF3 (NGG1p interacting factor 3)-like |
|
|
| 81 | c5b4bB_ |
|
not modelled |
74.3 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:udp-2,3-diacylglucosamine hydrolase;
PDBTitle: crystal structure of lpxh with lipid x in spacegroup c2
|
|
|
| 82 | d2fywa1 |
|
not modelled |
74.2 |
22 |
Fold:NIF3 (NGG1p interacting factor 3)-like
Superfamily:NIF3 (NGG1p interacting factor 3)-like
Family:NIF3 (NGG1p interacting factor 3)-like |
|
|
| 83 | c5w8mD_ |
|
not modelled |
73.9 |
12 |
PDB header:endocytosis
Chain: D: PDB Molecule:vacuolar protein sorting-associated protein 29;
PDBTitle: crystal structure of chaetomium thermophilum vps29
|
|
|
| 84 | d2a22a1 |
|
not modelled |
73.7 |
15 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:YfcE-like |
|
|
| 85 | c5ks8D_ |
|
not modelled |
73.2 |
17 |
PDB header:ligase
Chain: D: PDB Molecule:pyruvate carboxylase subunit beta;
PDBTitle: crystal structure of two-subunit pyruvate carboxylase from2 methylobacillus flagellatus
|
|
|
| 86 | c2dwuA_ |
|
not modelled |
73.2 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase isoform race1 from bacillus2 anthracis
|
|
|
| 87 | c3khjE_ |
|
not modelled |
72.9 |
17 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:inosine-5-monophosphate dehydrogenase;
PDBTitle: c. parvum inosine monophosphate dehydrogenase bound by inhibitor c64
|
|
|
| 88 | d1i60a_ |
|
not modelled |
71.4 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
|
|
| 89 | c4iwmD_ |
|
not modelled |
71.2 |
19 |
PDB header:unknown function
Chain: D: PDB Molecule:upf0135 protein mj0927;
PDBTitle: crystal structure of the conserved hypothetical protein mj0927 from2 methanocaldococcus jannaschii (in p21 form)
|
|
|
| 90 | d1h1ya_ |
|
not modelled |
71.0 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
|
|
| 91 | c3l9cA_ |
|
not modelled |
70.8 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: the crystal structure of smu.777 from streptococcus mutans ua159
|
|
|
| 92 | d1qhwa_ |
|
not modelled |
68.5 |
12 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Purple acid phosphatase-like |
|
|
| 93 | c1qhwA_ |
|
not modelled |
68.5 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein (purple acid phosphatase);
PDBTitle: purple acid phosphatase from rat bone
|
|
|
| 94 | c2y8kA_ |
|
not modelled |
68.4 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:carbohydrate binding family 6;
PDBTitle: structure of ctgh5-cbm6, an arabinoxylan-specific xylanase.
|
|
|
| 95 | c5tnvA_ |
|
not modelled |
67.8 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:ap endonuclease, family protein 2;
PDBTitle: crystal structure of a xylose isomerase-like tim barrel protein from2 mycobacterium smegmatis in complex with magnesium
|
|
|
| 96 | d1nmpa_ |
|
not modelled |
67.6 |
19 |
Fold:NIF3 (NGG1p interacting factor 3)-like
Superfamily:NIF3 (NGG1p interacting factor 3)-like
Family:NIF3 (NGG1p interacting factor 3)-like |
|
|
| 97 | c5zfsA_ |
|
not modelled |
67.5 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:d-allulose-3-epimerase;
PDBTitle: crystal structure of arthrobacter globiformis m30 sugar epimerase2 which can produce d-allulose from d-fructose
|
|
|
| 98 | d2q02a1 |
|
not modelled |
67.4 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
|
|
| 99 | d3d03a1 |
|
not modelled |
66.8 |
10 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:GpdQ-like |
|
|
| 100 | c5wlyA_ |
|
not modelled |
66.6 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:udp-2,3-diacylglucosamine hydrolase;
PDBTitle: e. coli lpxh- 8 mutations
|
|
|
| 101 | c3thdD_ |
|
not modelled |
66.5 |
13 |
PDB header:hydrolase
Chain: D: PDB Molecule:beta-galactosidase;
PDBTitle: crystal structure of human beta-galactosidase in complex with 1-2 deoxygalactonojirimycin
|
|
|
| 102 | d1w0ma_ |
|
not modelled |
66.1 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 103 | c2bmbA_ |
|
not modelled |
65.8 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:folic acid synthesis protein fol1;
PDBTitle: x-ray structure of the bifunctional 6-hydroxymethyl-7,8-2 dihydroxypterin pyrophosphokinase dihydropteroate synthase from3 saccharomyces cerevisiae
|
|
|
| 104 | c5k8kA_ |
|
not modelled |
65.8 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:udp-2,3-diacylglucosamine hydrolase;
PDBTitle: structure of the haemophilus influenzae lpxh-lipid x complex
|
|
|
| 105 | c2kknA_ |
|
not modelled |
65.1 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of themotoga maritima protein tm1076:2 northeast structural genomics consortium target vt57
|
|
|
| 106 | c3rxyA_ |
|
not modelled |
64.9 |
40 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:nif3 protein;
PDBTitle: crystal structure of nif3 superfamily protein from sphaerobacter2 thermophilus
|
|
|
| 107 | c2o7qA_ |
|
not modelled |
64.9 |
22 |
PDB header:oxidoreductase,transferase
Chain: A: PDB Molecule:bifunctional 3-dehydroquinate dehydratase/shikimate
PDBTitle: crystal structure of the a. thaliana dhq-dehydroshikimate-sdh-2 shikimate-nadp(h)
|
|
|
| 108 | d1b4ub_ |
|
not modelled |
64.8 |
13 |
Fold:Phosphorylase/hydrolase-like
Superfamily:LigB-like
Family:LigB-like |
|
|
| 109 | c5ks8F_ |
|
not modelled |
64.3 |
17 |
PDB header:ligase
Chain: F: PDB Molecule:pyruvate carboxylase subunit beta;
PDBTitle: crystal structure of two-subunit pyruvate carboxylase from2 methylobacillus flagellatus
|
|
|
| 110 | c3vylB_ |
|
not modelled |
64.0 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:l-ribulose 3-epimerase;
PDBTitle: structure of l-ribulose 3-epimerase
|
|
|
| 111 | d1hg3a_ |
|
not modelled |
63.1 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 112 | c5ijwA_ |
|
not modelled |
62.9 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: glutamate racemase (muri) from mycobacterium smegmatis with bound d-2 glutamate, 1.8 angstrom resolution, x-ray diffraction
|
|
|
| 113 | c2yybA_ |
|
not modelled |
62.2 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ttha1606;
PDBTitle: crystal structure of ttha1606 from thermus thermophilus hb8
|
|
|
| 114 | c6mp2B_ |
|
not modelled |
62.2 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:blman5b;
PDBTitle: crystal structure of blman5b solved by siras
|
|
|
| 115 | d1nvma2 |
|
not modelled |
62.1 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
|
|
| 116 | c1nvmG_ |
|
not modelled |
62.1 |
11 |
PDB header:lyase/oxidoreductase
Chain: G: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
|
|
|
| 117 | c2oczA_ |
|
not modelled |
61.2 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: the structure of a putative 3-dehydroquinate dehydratase from2 streptococcus pyogenes.
|
|
|
| 118 | c2nx9B_ |
|
not modelled |
61.2 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:oxaloacetate decarboxylase 2, subunit alpha;
PDBTitle: crystal structure of the carboxyltransferase domain of the2 oxaloacetate decarboxylase na+ pump from vibrio cholerae
|
|
|
| 119 | c3ijpA_ |
|
not modelled |
60.4 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: crystal structure of dihydrodipicolinate reductase from bartonella2 henselae at 2.0a resolution
|
|
|
| 120 | c2gzmB_ |
|
not modelled |
59.9 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of the glutamate racemase from bacillus anthracis
|
|
|