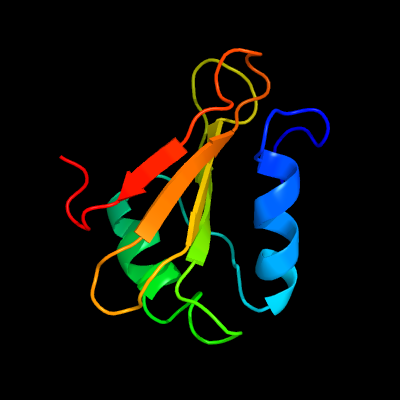
1 c2kgyA_
100.0
84
PDB header: immune systemChain: A: PDB Molecule: possible exported protein;PDBTitle: solution structure of rv0603 protein from mycobacterium2 tuberculosis h37rv
2 c4exrA_
74.9
18
PDB header: unknown functionChain: A: PDB Molecule: putative lipoprotein;PDBTitle: crystal structure of a putative lipoprotein (cd1622) from clostridium2 difficile 630 at 1.85 a resolution
3 c2vldA_
58.6
20
PDB header: hydrolaseChain: A: PDB Molecule: endonuclease nucs;PDBTitle: crystal structure of a repair endonuclease from pyrococcus abyssi
4 c5gkeB_
57.0
25
PDB header: hydrolase/dnaChain: B: PDB Molecule: endonuclease endoms;PDBTitle: structure of endoms-dsdna1 complex
5 c3riqA_
21.5
37
PDB header: viral proteinChain: A: PDB Molecule: tailspike protein;PDBTitle: siphovirus 9na tailspike receptor binding domain
6 c3hd4A_
19.9
29
PDB header: viral proteinChain: A: PDB Molecule: nucleoprotein;PDBTitle: mhv nucleocapsid protein ntd
7 d1s21a_
18.3
31
Fold: ADP-ribosylationSuperfamily: ADP-ribosylationFamily: AvrPphF ORF2, a type III effector
8 c1s21A_
18.3
31
PDB header: chaperoneChain: A: PDB Molecule: orf2;PDBTitle: crystal structure of avrpphf orf2, a type iii effector from p.2 syringae
9 d2cjoa_
13.6
37
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related
10 c6hosB_
13.6
28
PDB header: cell adhesionChain: B: PDB Molecule: ba75_04148t0;PDBTitle: structure of the kpflo2 adhesin domain in complex with glycerol
11 d3duea1
13.2
17
Fold: BLIP-likeSuperfamily: BT0923-likeFamily: BT0923-like
12 d2gyc31
11.8
64
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domainSuperfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domainFamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
13 c4v194_
11.6
29
PDB header: ribosomeChain: 4: PDB Molecule: mitoribosomal protein bl31m, mrpl55;PDBTitle: structure of the large subunit of the mammalian mitoribosome, part 12 of 2
14 c5n76C_
10.6
54
PDB header: nickel-binding proteinChain: C: PDB Molecule: coot;PDBTitle: crystal structure of the apo-form of the co dehydrogenase accessory2 protein coot from rhodospirillum rubrum
15 d2d7na1
10.5
9
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)
16 c5e4rA_
9.4
28
PDB header: oxidoreductaseChain: A: PDB Molecule: ketol-acid reductoisomerase;PDBTitle: crystal structure of domain-duplicated synthetic class ii ketol-acid2 reductoisomerase 2ia_kari-dd
17 c5xcxB_
9.3
22
PDB header: immune systemChain: B: PDB Molecule: vl-sarah(s37c) chimera;PDBTitle: crystal structure of ts2/16 fv-clasp fragment
18 d1dlja2
8.9
22
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain
19 d1v2ya_
8.5
29
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ubiquitin-related
20 c3d8tB_
7.2
0
PDB header: lyaseChain: B: PDB Molecule: uroporphyrinogen-iii synthase;PDBTitle: thermus thermophilus uroporphyrinogen iii synthase
21 c3witA_
not modelled
7.0
50
PDB header: structural proteinChain: A: PDB Molecule: putative vgr protein;PDBTitle: crystal structure of the c-terminal region of vgrg1 from e. coli o1572 edl933
22 c2e12B_
not modelled
7.0
31
PDB header: translationChain: B: PDB Molecule: hypothetical protein xcc3642;PDBTitle: the crystal structure of xc5848 from xanthomonas campestris2 adopting a novel variant of sm-like motif
23 c5kdvA_
not modelled
6.7
22
PDB header: hydrolaseChain: A: PDB Molecule: metallopeptidase;PDBTitle: impa metallopeptidase from pseudomonas aeruginosa
24 d1np3a2
not modelled
6.7
30
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain
25 c2qtzA_
not modelled
6.6
46
PDB header: oxidoreductaseChain: A: PDB Molecule: methionine synthase reductase;PDBTitle: crystal structure of the nadp+-bound fad-containing fnr-like module of2 human methionine synthase reductase
26 d1dd3a1
not modelled
6.3
39
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domainSuperfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domainFamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
27 c5cb9A_
not modelled
5.9
30
PDB header: lyaseChain: A: PDB Molecule: glyoxalase/bleomycin resistance protein/dioxygenase;PDBTitle: crystal structure of c-as lyase with mercaptoethonal
28 c4oleD_
not modelled
5.7
38
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: next to brca1 gene 1 protein;PDBTitle: crystal structure of a neighbor of brca1 gene 1 (nbr1) from homo2 sapiens at 2.52 a resolution
29 c1f20A_
not modelled
5.7
46
PDB header: oxidoreductaseChain: A: PDB Molecule: nitric-oxide synthase;PDBTitle: crystal structure of rat neuronal nitric-oxide synthase fad/nadp+2 domain at 1.9a resolution.
30 d3elga1
not modelled
5.7
17
Fold: BLIP-likeSuperfamily: BT0923-likeFamily: BT0923-like
31 c2m7yA_
not modelled
5.7
67
PDB header: viral proteinChain: A: PDB Molecule: leader peptide;PDBTitle: the mengovirus leader protein
32 c3qftA_
not modelled
5.7
38
PDB header: oxidoreductaseChain: A: PDB Molecule: nadph--cytochrome p450 reductase;PDBTitle: crystal structure of nadph-cytochrome p450 reductase (fad/nadph domain2 and r457h mutant)
33 c6j7aB_
not modelled
5.5
46
PDB header: oxidoreductaseChain: B: PDB Molecule: heme oxygenase 1,nadph--cytochrome p450 reductase;PDBTitle: fusion protein of heme oxygenase-1 and nadph cytochrome p450 reductase2 (17aa)
34 c5omtA_
not modelled
5.5
44
PDB header: hydrolaseChain: A: PDB Molecule: nucb;PDBTitle: endonuclease nucb
35 d1jb9a2
not modelled
5.4
38
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: Reductases
36 c1yrlD_
not modelled
5.3
30
PDB header: oxidoreductaseChain: D: PDB Molecule: ketol-acid reductoisomerase;PDBTitle: escherichia coli ketol-acid reductoisomerase
37 c5igrA_
not modelled
5.2
26
PDB header: transferase/antibioticChain: A: PDB Molecule: macrolide 2'-phosphotransferase;PDBTitle: macrolide 2'-phosphotransferase type i - complex with gdp and2 oleandomycin
38 c5wecB_
not modelled
5.2
36
PDB header: protein transportChain: B: PDB Molecule: lipoprotein;PDBTitle: structure of an alternative pilotin from the type ii secretion system2 of vibrio cholerae
39 c4dqkA_
not modelled
5.1
38
PDB header: oxidoreductaseChain: A: PDB Molecule: bifunctional p-450/nadph-p450 reductase;PDBTitle: crystal structure of the fad binding domain of cytochrome p450 bm3