1 c5in3A_
100.0
29
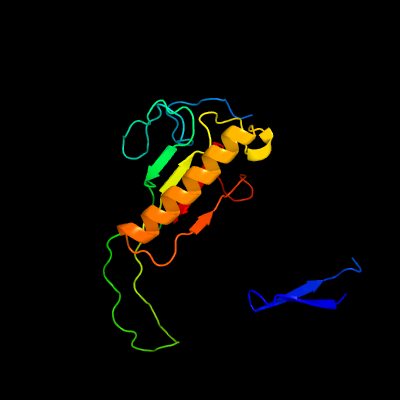
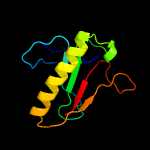
PDB header: transferaseChain: A: PDB Molecule: galactose-1-phosphate uridylyltransferase;PDBTitle: crystal structure of glucose-1-phosphate bound nucleotidylated human2 galactose-1-phosphate uridylyltransferase
2 c1gupC_
100.0
26
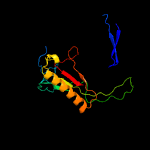
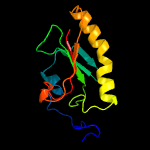
PDB header: nucleotidyltransferaseChain: C: PDB Molecule: galactose-1-phosphate uridylyltransferase;PDBTitle: structure of nucleotidyltransferase complexed with udp-2 galactose
3 d1guqa1
100.0
27
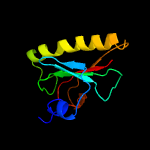
Fold: HIT-likeSuperfamily: HIT-likeFamily: Hexose-1-phosphate uridylyltransferase
4 c1zwjA_
100.0
23
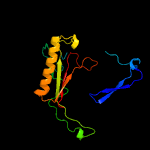
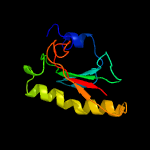
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative galactose-1-phosphate uridyl transferase;PDBTitle: x-ray structure of galt-like protein from arabidopsis thaliana2 at5g18200
5 d1z84a1
100.0
23
Fold: HIT-likeSuperfamily: HIT-likeFamily: Hexose-1-phosphate uridylyltransferase
6 c4qvuA_
99.9
19
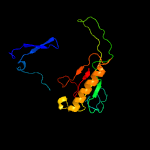
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a duf4931 family protein (bce0241) from bacillus2 cereus atcc 10987 at 2.65 a resolution
7 c4i5wA_
98.4
19
PDB header: transferaseChain: A: PDB Molecule: 5',5'''-p-1,p-4-tetraphosphate phosphorylase 2;PDBTitle: crystal structure of yeast ap4a phosphorylase apa2 in complex with amp
8 c5cs2A_
98.0
16
PDB header: hydrolaseChain: A: PDB Molecule: histidine triad protein;PDBTitle: crystal structure of plasmodium falciparum diadenosine triphosphate2 hydrolase in complex with cyclomarin a
9 c3ksvA_
97.8
18
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: hypothetical protein from leishmania major
10 c3anoA_
97.6
17
PDB header: transferaseChain: A: PDB Molecule: ap-4-a phosphorylase;PDBTitle: crystal structure of a novel diadenosine 5',5'''-p1,p4-tetraphosphate2 phosphorylase from mycobacterium tuberculosis h37rv
11 c2eo4A_
97.4
13
PDB header: hydrolaseChain: A: PDB Molecule: 150aa long hypothetical histidine triad nucleotide-bindingPDBTitle: crystal structure of hypothetical histidine triad nucleotide-binding2 protein st2152 from sulfolobus tokodaii strain7
12 d1emsa1
97.3
19
Fold: HIT-likeSuperfamily: HIT-likeFamily: HIT (HINT, histidine triad) family of protein kinase-interacting proteins
13 c3l7xA_
97.2
16
PDB header: cell cycleChain: A: PDB Molecule: putative hit-like protein involved in cell-cyclePDBTitle: the crystal structure of smu.412c from streptococcus mutans ua159
14 d1y23a_
97.2
17
Fold: HIT-likeSuperfamily: HIT-likeFamily: HIT (HINT, histidine triad) family of protein kinase-interacting proteins
15 c3imiB_
97.2
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hit family protein;PDBTitle: 2.01 angstrom resolution crystal structure of a hit family protein2 from bacillus anthracis str. 'ames ancestor'
16 d1fita_
97.2
17
Fold: HIT-likeSuperfamily: HIT-likeFamily: HIT (HINT, histidine triad) family of protein kinase-interacting proteins
17 c3lb5B_
97.1
13
PDB header: cell cycleChain: B: PDB Molecule: hit-like protein involved in cell-cycle regulation;PDBTitle: crystal structure of hit-like protein involved in cell-cycle2 regulation from bartonella henselae with unknown ligand
18 c3o0mB_
97.0
13
PDB header: hydrolaseChain: B: PDB Molecule: hit family protein;PDBTitle: crystal structure of a zn-bound histidine triad family protein from2 mycobacterium smegmatis
19 c3oj7A_
97.0
13
PDB header: metal binding proteinChain: A: PDB Molecule: putative histidine triad family protein;PDBTitle: crystal structure of a histidine triad family protein from entamoeba2 histolytica, bound to sulfate
20 c6iq1A_
96.8
14
PDB header: hydrolaseChain: A: PDB Molecule: adenosine 5'-monophosphoramidase;PDBTitle: crystal structure of histidine triad nucleotide-binding protein from2 candida albicans
21 c1xquA_
not modelled
96.7
16
PDB header: hydrolaseChain: A: PDB Molecule: hit family hydrolase;PDBTitle: hit family hydrolase from clostridium thermocellum cth-393
22 d1xqua_
not modelled
96.7
16
Fold: HIT-likeSuperfamily: HIT-likeFamily: HIT (HINT, histidine triad) family of protein kinase-interacting proteins
23 c1emsB_
not modelled
96.7
20
PDB header: antitumor proteinChain: B: PDB Molecule: nit-fragile histidine triad fusion protein;PDBTitle: crystal structure of the c. elegans nitfhit protein
24 d1kpfa_
not modelled
96.6
11
Fold: HIT-likeSuperfamily: HIT-likeFamily: HIT (HINT, histidine triad) family of protein kinase-interacting proteins
25 d1rzya_
not modelled
96.6
13
Fold: HIT-likeSuperfamily: HIT-likeFamily: HIT (HINT, histidine triad) family of protein kinase-interacting proteins
26 c4eguA_
not modelled
96.6
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: histidine triad (hit) protein;PDBTitle: 0.95a resolution structure of a histidine triad protein from2 clostridium difficile
27 c6d6jB_
not modelled
96.6
12
PDB header: hydrolaseChain: B: PDB Molecule: hit family hydrolase;PDBTitle: crystal structure of hit family hydrolase from legionella pneumophila2 philadelphia 1
28 c4incA_
not modelled
96.2
11
PDB header: hydrolaseChain: A: PDB Molecule: histidine triad nucleotide-binding protein 2,PDBTitle: human histidine triad nucleotide binding protein 2
29 d1z84a2
not modelled
95.8
15
Fold: HIT-likeSuperfamily: HIT-likeFamily: Hexose-1-phosphate uridylyltransferase
30 d2oika1
not modelled
95.8
17
Fold: HIT-likeSuperfamily: HIT-likeFamily: HIT (HINT, histidine triad) family of protein kinase-interacting proteins
31 c4q61J_
not modelled
95.6
10
PDB header: cell cycleChain: J: PDB Molecule: uncharacterized hit-like protein hp_0404;PDBTitle: hit like protein from helicobacter pylori 26695
32 c3n1tE_
not modelled
95.2
9
PDB header: hydrolaseChain: E: PDB Molecule: hit-like protein hint;PDBTitle: crystal structure of the h101a mutant echint gmp complex
33 c3r6fA_
not modelled
94.8
16
PDB header: hydrolaseChain: A: PDB Molecule: hit family protein;PDBTitle: crystal structure of a zinc-containing hit family protein from2 encephalitozoon cuniculi
34 c3p0tB_
not modelled
94.6
16
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an hit-like protein from mycobacterium2 paratuberculosis
35 c3jb9c_
not modelled
93.1
16
PDB header: rna binding protein/rnaChain: C: PDB Molecule: u5 snrna;PDBTitle: cryo-em structure of the yeast spliceosome at 3.6 angstrom resolution
36 c3i4sB_
not modelled
90.2
8
PDB header: hydrolaseChain: B: PDB Molecule: histidine triad protein;PDBTitle: crystal structure of histidine triad protein blr8122 from2 bradyrhizobium japonicum
37 d1guqa2
not modelled
89.6
13
Fold: HIT-likeSuperfamily: HIT-likeFamily: Hexose-1-phosphate uridylyltransferase
38 c3oheA_
not modelled
88.5
8
PDB header: hydrolaseChain: A: PDB Molecule: histidine triad (hit) protein;PDBTitle: crystal structure of a histidine triad protein (maqu_1709) from2 marinobacter aquaeolei vt8 at 1.20 a resolution
39 c3i24B_
not modelled
85.8
5
PDB header: hydrolaseChain: B: PDB Molecule: hit family hydrolase;PDBTitle: crystal structure of a hit family hydrolase protein from vibrio2 fischeri. northeast structural genomics consortium target id vfr176
40 c6id1U_
not modelled
83.3
13
PDB header: splicingChain: U: PDB Molecule: cwf19-like protein 2;PDBTitle: cryo-em structure of a human intron lariat spliceosome after prp432 loaded (ils2 complex) at 2.9 angstrom resolution
41 c3nrdB_
not modelled
80.3
10
PDB header: nucleotide binding proteinChain: B: PDB Molecule: histidine triad (hit) protein;PDBTitle: crystal structure of a histidine triad (hit) protein (smc02904) from2 sinorhizobium meliloti 1021 at 2.06 a resolution
42 c5sxyA_
not modelled
50.6
41
PDB header: chaperoneChain: A: PDB Molecule: bifunctional coenzyme pqq synthesis protein c/d;PDBTitle: the solution nmr structure for the pqqd truncation of methylobacterium2 extorquens pqqcd representing a functional and stand-alone3 ribosomally synthesized and post-translational modified (ripp)4 recognition element (rre)
43 c4ndgB_
not modelled
46.4
16
PDB header: dna binding protein/rna/dnaChain: B: PDB Molecule: aprataxin;PDBTitle: human aprataxin (aptx) bound to rna-dna and zn - adenosine vanadate2 transition state mimic complex
44 d1vpra1
not modelled
38.4
20
Fold: LipocalinsSuperfamily: LipocalinsFamily: Dinoflagellate luciferase repeat
45 c3g2bA_
not modelled
36.6
41
PDB header: biosynthetic proteinChain: A: PDB Molecule: coenzyme pqq synthesis protein d;PDBTitle: crystal structure of pqqd from xanthomonas campestris
46 c2iqcA_
not modelled
26.7
46
PDB header: protein bindingChain: A: PDB Molecule: fanconi anemia group f protein;PDBTitle: crystal structure of human fancf protein that functions in2 the assembly of a dna damage signaling complex
47 c1t3bA_
not modelled
26.1
14
PDB header: isomeraseChain: A: PDB Molecule: thiol:disulfide interchange protein dsbc;PDBTitle: x-ray structure of dsbc from haemophilus influenzae
48 c3pa8A_
not modelled
24.7
21
PDB header: toxin/peptide inhibitorChain: A: PDB Molecule: toxin b;PDBTitle: structure of the c. difficile tcdb cysteine protease domain in complex2 with a peptide inhibitor
49 d1igna2
not modelled
24.1
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: DNA-binding domain of rap1
50 c3j6vR_
not modelled
22.7
23
PDB header: ribosomeChain: R: PDB Molecule: 28s ribosomal protein s18a, mitochondrial;PDBTitle: cryo-em structure of the small subunit of the mammalian mitochondrial2 ribosome
51 d1ea9c2
not modelled
21.1
24
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain
52 c4acvA_
not modelled
20.4
40
PDB header: immune systemChain: A: PDB Molecule: prophage lambdalm01, antigen b;PDBTitle: listeria monocytogenes antigen b
53 c4lh9A_
not modelled
19.9
12
PDB header: transcriptionChain: A: PDB Molecule: heterocyst differentiation control protein;PDBTitle: crystal structure of the refolded hood domain (asp256-gly295) of hetr
54 c5aj3R_
not modelled
19.8
43
PDB header: ribosomeChain: R: PDB Molecule: mitoribosomal protein bs18m, mrps18c;PDBTitle: structure of the small subunit of the mammalian mitoribosome
55 d1u0la1
not modelled
19.8
20
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
56 d1t9ha1
not modelled
18.3
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
57 c5xmcA_
not modelled
18.2
19
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase itchy;PDBTitle: crystal structure of the auto-inhibited nedd4 family e3 ligase itch
58 d1w2za2
not modelled
16.0
6
Fold: Cystatin-likeSuperfamily: Amine oxidase N-terminal regionFamily: Amine oxidase N-terminal region
59 c5a3gA_
not modelled
16.0
17
PDB header: viral proteinChain: A: PDB Molecule: m50;PDBTitle: structure of herpesvirus nuclear egress complex subunit m50
60 c3t4cD_
not modelled
15.7
6
PDB header: transferaseChain: D: PDB Molecule: 2-dehydro-3-deoxyphosphooctonate aldolase 1;PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia ambifaria
61 c1rfoC_
not modelled
15.4
38
PDB header: viral proteinChain: C: PDB Molecule: whisker antigen control protein;PDBTitle: trimeric foldon of the t4 phagehead fibritin
62 c2he3A_
not modelled
15.0
21
PDB header: oxidoreductaseChain: A: PDB Molecule: glutathione peroxidase 2;PDBTitle: crystal structure of the selenocysteine to cysteine mutant of human2 glutathionine peroxidase 2 (gpx2)
63 d1n6ja_
not modelled
14.6
11
Fold: SRF-likeSuperfamily: SRF-likeFamily: SRF-like
64 c5yirG_
not modelled
14.6
50
PDB header: protein bindingChain: G: PDB Molecule: ankyrin-2;PDBTitle: crystal structure of ankb lir/gabarap complex
65 c3w6gP_
not modelled
14.6
35
PDB header: oxidoreductaseChain: P: PDB Molecule: probable peroxiredoxin;PDBTitle: structure of peroxiredoxin from anaerobic hyperthermophilic archaeon2 pyrococcus horikoshii
66 c5n6yC_
not modelled
14.3
4
PDB header: oxidoreductaseChain: C: PDB Molecule: nitrogenase vanadium-iron protein delta chain;PDBTitle: azotobacter vinelandii vanadium nitrogenase
67 c3epsB_
not modelled
14.2
15
PDB header: transferase, hydrolaseChain: B: PDB Molecule: isocitrate dehydrogenase kinase/phosphatase;PDBTitle: the crystal structure of isocitrate dehydrogenase kinase/phosphatase2 from e. coli
68 d1z8ma1
not modelled
14.1
20
Fold: RelE-likeSuperfamily: RelE-likeFamily: RelE-like
69 d2lhba_
not modelled
14.1
24
Fold: Globin-likeSuperfamily: Globin-likeFamily: Globins
70 c2rilA_
not modelled
13.9
11
PDB header: oxidoreductaseChain: A: PDB Molecule: antibiotic biosynthesis monooxygenase;PDBTitle: crystal structure of a putative monooxygenase (yp_001095275.1) from2 shewanella loihica pv-4 at 1.26 a resolution
71 c4uhpB_
not modelled
13.8
24
PDB header: hydrolaseChain: B: PDB Molecule: bacteriocin immunity protein;PDBTitle: crystal structure of the pyocin ap41 dnase-immunity complex
72 c3znuI_
not modelled
13.3
14
PDB header: lyaseChain: I: PDB Molecule: 5-chloromuconolactone dehalogenase;PDBTitle: crystal structure of clcf in crystal form 2
73 c2bc4C_
not modelled
13.1
40
PDB header: immune systemChain: C: PDB Molecule: hla class ii histocompatibility antigen, dm alpha chain;PDBTitle: crystal structure of hla-dm
74 d1egwa_
not modelled
13.0
11
Fold: SRF-likeSuperfamily: SRF-likeFamily: SRF-like
75 c1f3jD_
not modelled
12.8
40
PDB header: immune systemChain: D: PDB Molecule: h-2 class ii histocompatibility antigen;PDBTitle: histocompatibility antigen i-ag7
76 d1ocya_
not modelled
12.7
23
Fold: Receptor-binding domain of short tail fibre protein gp12Superfamily: Receptor-binding domain of short tail fibre protein gp12Family: Receptor-binding domain of short tail fibre protein gp12
77 c4ynlB_
not modelled
12.6
12
PDB header: transcriptionChain: B: PDB Molecule: heterocyst differentiation control protein;PDBTitle: crystal structure of the hood domain of anabaena hetr in complex with2 the hexapeptide ergsgr derived from pats
78 d1d6za2
not modelled
12.4
6
Fold: Cystatin-likeSuperfamily: Amine oxidase N-terminal regionFamily: Amine oxidase N-terminal region
79 c3fq6A_
not modelled
12.1
29
PDB header: transferaseChain: A: PDB Molecule: methyltransferase;PDBTitle: the crystal structure of a methyltransferase domain from bacteroides2 thetaiotaomicron vpi
80 d1kwaa_
not modelled
11.7
32
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain
81 d2f8aa1
not modelled
11.7
11
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like
82 c3mlcC_
not modelled
11.6
13
PDB header: isomeraseChain: C: PDB Molecule: fg41 malonate semialdehyde decarboxylase;PDBTitle: crystal structure of fg41msad inactivated by 3-chloropropiolate
83 c2oruA_
not modelled
11.6
50
PDB header: de novo proteinChain: A: PDB Molecule: xtz1-peptide;PDBTitle: solution structure of xtz1-peptide, a beta-hairpin peptide2 with a structured extension
84 c1vluB_
not modelled
11.6
19
PDB header: oxidoreductaseChain: B: PDB Molecule: gamma-glutamyl phosphate reductase;PDBTitle: crystal structure of gamma-glutamyl phosphate reductase (yor323c) from2 saccharomyces cerevisiae at 2.40 a resolution
85 d1ua7a1
not modelled
11.5
29
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain
86 d1zaka2
not modelled
11.4
50
Fold: Rubredoxin-likeSuperfamily: Microbial and mitochondrial ADK, insert "zinc finger" domainFamily: Microbial and mitochondrial ADK, insert "zinc finger" domain
87 c4qglA_
not modelled
11.3
12
PDB header: oxidoreductaseChain: A: PDB Molecule: acireductone dioxygenase;PDBTitle: acireductone dioxygenase from bacillus anthracis with three cadmium2 ions
88 c1ekmC_
not modelled
11.3
23
PDB header: oxidoreductaseChain: C: PDB Molecule: copper amine oxidase;PDBTitle: crystal structure at 2.5 a resolution of zinc-substituted2 copper amine oxidase of hansenula polymorpha expressed in3 escherichia coli
89 c5yisC_
not modelled
11.1
36
PDB header: protein bindingChain: C: PDB Molecule: ankyrin-2;PDBTitle: crystal structure of ankb lir/lc3b complex
90 d1nqua_
not modelled
11.0
41
Fold: Lumazine synthaseSuperfamily: Lumazine synthaseFamily: Lumazine synthase
91 c3sdsA_
not modelled
11.0
18
PDB header: transferaseChain: A: PDB Molecule: ornithine carbamoyltransferase, mitochondrial;PDBTitle: crystal structure of a mitochondrial ornithine carbamoyltransferase2 from coccidioides immitis
92 c3nbbC_
not modelled
11.0
23
PDB header: oxidoreductaseChain: C: PDB Molecule: peroxisomal primary amine oxidase;PDBTitle: crystal structure of mutant y305f expressed in e. coli in the copper2 amine oxidase from hansenula polymorpha
93 d1w6ga2
not modelled
10.9
14
Fold: Cystatin-likeSuperfamily: Amine oxidase N-terminal regionFamily: Amine oxidase N-terminal region
94 c2elpA_
not modelled
10.8
57
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 13th c2h2 zinc finger of human2 zinc finger protein 406
95 c4xruE_
not modelled
10.8
11
PDB header: protein bindingChain: E: PDB Molecule: rnl;PDBTitle: structure of pnkp1/rnl/hen1 complex
96 c3ho6B_
not modelled
10.6
17
PDB header: toxinChain: B: PDB Molecule: toxin a;PDBTitle: structure-function analysis of inositol hexakisphosphate-2 induced autoprocessing in clostridium difficile toxin a
97 c1yuzB_
not modelled
10.5
38
PDB header: oxidoreductaseChain: B: PDB Molecule: nigerythrin;PDBTitle: partially reduced state of nigerythrin
98 c4q2uH_
not modelled
10.5
20
PDB header: toxin/toxin repressorChain: H: PDB Molecule: mrna interferase yafq;PDBTitle: crystal structure of the e. coli dinj-yafq toxin-antitoxin complex
99 c6jy5B_
not modelled
10.4
31
PDB header: structural proteinChain: B: PDB Molecule: unidentified carboxysome polypeptide;PDBTitle: structure of csos4b from halothiobacillus neapolitanus