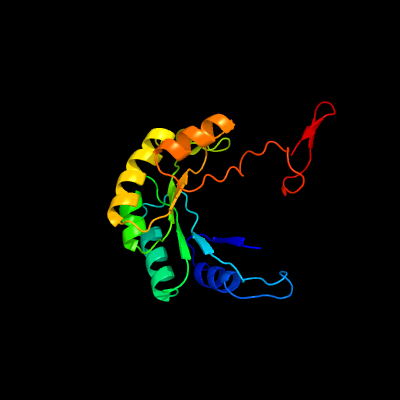
1 c1zfjA_
79.2
15
PDB header: oxidoreductaseChain: A: PDB Molecule: inosine monophosphate dehydrogenase;PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
2 d1rq2a2
62.7
16
Fold: Bacillus chorismate mutase-likeSuperfamily: Tubulin C-terminal domain-likeFamily: Tubulin, C-terminal domain
3 d2vapa2
58.9
21
Fold: Bacillus chorismate mutase-likeSuperfamily: Tubulin C-terminal domain-likeFamily: Tubulin, C-terminal domain
4 d1w5fa2
55.4
17
Fold: Bacillus chorismate mutase-likeSuperfamily: Tubulin C-terminal domain-likeFamily: Tubulin, C-terminal domain
5 c2zauB_
54.8
18
PDB header: transferaseChain: B: PDB Molecule: selenide, water dikinase;PDBTitle: crystal structure of an n-terminally truncated2 selenophosphate synthetase from aquifex aeolicus
6 d1dbfa_
51.0
13
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: Chorismate mutase
7 d1vk3a2
41.6
13
Fold: Bacillus chorismate mutase-likeSuperfamily: PurM N-terminal domain-likeFamily: PurM N-terminal domain-like
8 d1ufya_
38.3
13
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: Chorismate mutase
9 d1ofua2
38.2
16
Fold: Bacillus chorismate mutase-likeSuperfamily: Tubulin C-terminal domain-likeFamily: Tubulin, C-terminal domain
10 c4z87B_
36.7
16
PDB header: oxidoreductaseChain: B: PDB Molecule: inosine-5'-monophosphate dehydrogenase;PDBTitle: structure of the imp dehydrogenase from ashbya gossypii bound to gdp
11 c1xhoB_
34.7
8
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: chorismate mutase;PDBTitle: chorismate mutase from clostridium thermocellum cth-682
12 d1xhoa_
34.7
8
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: Chorismate mutase
13 c4r3uD_
34.1
2
PDB header: isomeraseChain: D: PDB Molecule: 2-hydroxyisobutyryl-coa mutase small subunit;PDBTitle: crystal structure of 2-hydroxyisobutyryl-coa mutase
14 c4ldaF_
33.9
6
PDB header: transcriptionChain: F: PDB Molecule: tadz;PDBTitle: crystal structure of a chey-like protein (tadz) from pseudomonas2 aeruginosa pao1 at 2.70 a resolution
15 c2xdqA_
32.5
13
PDB header: oxidoreductaseChain: A: PDB Molecule: light-independent protochlorophyllide reductase subunit n;PDBTitle: dark operative protochlorophyllide oxidoreductase (chln-chlb)2 complex
16 c2ayxA_
31.4
14
PDB header: transferaseChain: A: PDB Molecule: sensor kinase protein rcsc;PDBTitle: solution structure of the e.coli rcsc c-terminus (residues2 700-949) containing linker region and phosphoreceiver3 domain
17 c1vliA_
27.3
20
PDB header: biosynthetic proteinChain: A: PDB Molecule: spore coat polysaccharide biosynthesis protein spse;PDBTitle: crystal structure of spore coat polysaccharide biosynthesis protein2 spse (bsu37870) from bacillus subtilis at 2.38 a resolution
18 d2zaua1
26.7
18
Fold: Bacillus chorismate mutase-likeSuperfamily: PurM N-terminal domain-likeFamily: PurM N-terminal domain-like
19 c5uicA_
26.1
15
PDB header: transcriptionChain: A: PDB Molecule: two-component response regulator;PDBTitle: structure of the francisella response regulator receiver domain, qseb
20 d1qh8a_
25.0
16
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
21 c1ny5A_
not modelled
24.9
12
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: crystal structure of sigm54 activator (aaa+ atpase) in the inactive2 state
22 c3rqiA_
not modelled
24.0
10
PDB header: transcriptionChain: A: PDB Molecule: response regulator protein;PDBTitle: crystal structure of a response regulator protein from burkholderia2 pseudomallei with a phosphorylated aspartic acid, calcium ion and3 citrate
23 d1b74a1
not modelled
23.6
8
Fold: ATC-likeSuperfamily: Aspartate/glutamate racemaseFamily: Aspartate/glutamate racemase
24 d1m1na_
not modelled
23.1
11
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
25 d1qe5a_
not modelled
22.0
29
Fold: Phosphorylase/hydrolase-likeSuperfamily: Purine and uridine phosphorylasesFamily: Purine and uridine phosphorylases
26 c4k40B_
not modelled
22.0
18
PDB header: hydrolaseChain: B: PDB Molecule: gdsl-like lipase/acylhydrolase family protein;PDBTitle: peptidoglycan o-acetylesterase in action, 0 min
27 d1g2oa_
not modelled
22.0
33
Fold: Phosphorylase/hydrolase-likeSuperfamily: Purine and uridine phosphorylasesFamily: Purine and uridine phosphorylases
28 c2yxbA_
not modelled
21.9
6
PDB header: isomeraseChain: A: PDB Molecule: coenzyme b12-dependent mutase;PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
29 c6ekhY_
not modelled
21.3
22
PDB header: metal binding proteinChain: Y: PDB Molecule: chemotaxis protein chey;PDBTitle: crystal structure of activated chey from methanoccocus maripaludis
30 c5wlpA_
not modelled
21.0
13
PDB header: protein transportChain: A: PDB Molecule: autophagy-related protein 32;PDBTitle: solution structure of the pseudo-receiver domain of atg32
31 d1ys7a2
not modelled
20.8
18
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related
32 d3c9ua1
not modelled
20.4
14
Fold: Bacillus chorismate mutase-likeSuperfamily: PurM N-terminal domain-likeFamily: PurM N-terminal domain-like
33 c3bolB_
not modelled
19.2
10
PDB header: transferaseChain: B: PDB Molecule: 5-methyltetrahydrofolate s-homocysteinePDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
34 c5ep0A_
not modelled
18.8
16
PDB header: transcriptionChain: A: PDB Molecule: putative repressor protein luxo;PDBTitle: quorum-sensing signal integrator luxo - receiver+catalytic domains
35 c5n6yD_
not modelled
18.5
16
PDB header: oxidoreductaseChain: D: PDB Molecule: nitrogenase vanadium-iron protein alpha chain;PDBTitle: azotobacter vinelandii vanadium nitrogenase
36 c3stgA_
not modelled
18.0
16
PDB header: transferaseChain: A: PDB Molecule: 2-dehydro-3-deoxyphosphooctonate aldolase;PDBTitle: crystal structure of a58p, del(n59), and loop 7 truncated mutant of 3-2 deoxy-d-manno-octulosonate 8-phosphate synthase (kdo8ps) from3 neisseria meningitidis
37 c5lwkB_
not modelled
17.9
20
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulatory protein;PDBTitle: maer response regulator bound to beryllium trifluoride
38 c4qpjC_
not modelled
17.9
8
PDB header: signaling protein/dna binding proteinChain: C: PDB Molecule: cell cycle response regulator ctra;PDBTitle: 2.7 angstrom structure of a phosphotransferase in complex with a2 receiver domain
39 c4rndB_
not modelled
17.9
13
PDB header: hydrolaseChain: B: PDB Molecule: v-type proton atpase subunit f;PDBTitle: crystal structure of the subunit df-assembly of the eukaryotic v-2 atpase.
40 c3dzdA_
not modelled
17.4
14
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: crystal structure of sigma54 activator ntrc4 in the inactive2 state
41 c6ifhA_
not modelled
17.3
18
PDB header: transferaseChain: A: PDB Molecule: sporulation initiation phosphotransferase f;PDBTitle: unphosphorylated spo0f from paenisporosarcina sp. tg-14
42 c3hv2B_
not modelled
17.2
20
PDB header: signaling proteinChain: B: PDB Molecule: response regulator/hd domain protein;PDBTitle: crystal structure of signal receiver domain of hd domain-containing2 protein from pseudomonas fluorescens pf-5
43 d1qh8b_
not modelled
17.1
11
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
44 c3g8rA_
not modelled
16.8
9
PDB header: biosynthetic proteinChain: A: PDB Molecule: probable spore coat polysaccharide biosynthesis protein e;PDBTitle: crystal structure of putative spore coat polysaccharide biosynthesis2 protein e from chromobacterium violaceum atcc 12472
45 c3cu5B_
not modelled
16.2
10
PDB header: transcription regulatorChain: B: PDB Molecule: two component transcriptional regulator, arac family;PDBTitle: crystal structure of a two component transcriptional regulator arac2 from clostridium phytofermentans isdg
46 c1y80A_
not modelled
15.9
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: predicted cobalamin binding protein;PDBTitle: structure of a corrinoid (factor iiim)-binding protein from moorella2 thermoacetica
47 c4q7eA_
not modelled
15.8
16
PDB header: signaling proteinChain: A: PDB Molecule: response regulator of a two component regulatory system;PDBTitle: non-phosphorylated hemr receiver domain from leptospira biflexa
48 c1w59B_
not modelled
15.8
21
PDB header: cell divisionChain: B: PDB Molecule: cell division protein ftsz homolog 1;PDBTitle: ftsz dimer, empty (m. jannaschii)
49 d1vlia2
not modelled
15.5
22
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: NeuB-like
50 c5i4cA_
not modelled
15.4
8
PDB header: gene regulationChain: A: PDB Molecule: transcriptional regulatory protein rcsb;PDBTitle: crystal structure of non-phosphorylated receiver domain of the stress2 response regulator rcsb from escherichia coli
51 c2rjnA_
not modelled
15.3
16
PDB header: hydrolaseChain: A: PDB Molecule: response regulator receiver:metal-dependentPDBTitle: crystal structure of an uncharacterized protein q2bku2 from2 neptuniibacter caesariensis
52 c2ynmC_
not modelled
15.2
15
PDB header: oxidoreductaseChain: C: PDB Molecule: light-independent protochlorophyllide reductase subunit n;PDBTitle: structure of the adpxalf3-stabilized transition state of the2 nitrogenase-like dark-operative protochlorophyllide oxidoreductase3 complex from prochlorococcus marinus with its substrate4 protochlorophyllide a
53 d1mvoa_
not modelled
14.9
18
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related
54 c3outC_
not modelled
14.5
14
PDB header: isomeraseChain: C: PDB Molecule: glutamate racemase;PDBTitle: crystal structure of glutamate racemase from francisella tularensis2 subsp. tularensis schu s4 in complex with d-glutamate.
55 d1ny5a1
not modelled
14.2
12
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related
56 c3sz8D_
not modelled
14.2
15
PDB header: transferaseChain: D: PDB Molecule: 2-dehydro-3-deoxyphosphooctonate aldolase 2;PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia pseudomallei
57 d1m1nb_
not modelled
14.0
4
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
58 d1krwa_
not modelled
14.0
16
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related
59 c4e7pA_
not modelled
14.0
10
PDB header: transcription regulatorChain: A: PDB Molecule: response regulator;PDBTitle: crystal structure of receiver domain of putative narl family response2 regulator spr1814 from streptococcus pneumoniae in the presence of3 the phosphoryl analog beryllofluoride
60 d1peya_
not modelled
14.0
20
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related
61 c3pdiB_
not modelled
13.8
18
PDB header: protein bindingChain: B: PDB Molecule: nitrogenase mofe cofactor biosynthesis protein nifn;PDBTitle: precursor bound nifen
62 d1kgsa2
not modelled
13.8
8
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related
63 c2vxyA_
not modelled
13.5
16
PDB header: cell cycleChain: A: PDB Molecule: cell division protein ftsz;PDBTitle: the structure of ftsz from bacillus subtilis at 1.7a2 resolution
64 d1jbea_
not modelled
13.4
14
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related
65 c3b2nA_
not modelled
13.3
14
PDB header: transcriptionChain: A: PDB Molecule: uncharacterized protein q99uf4;PDBTitle: crystal structure of dna-binding response regulator, luxr family, from2 staphylococcus aureus
66 c1xuzA_
not modelled
12.7
17
PDB header: biosynthetic proteinChain: A: PDB Molecule: polysialic acid capsule biosynthesis protein siac;PDBTitle: crystal structure analysis of sialic acid synthase (neub)from2 neisseria meningitidis, bound to mn2+, phosphoenolpyruvate, and n-3 acetyl mannosaminitol
67 c5m7nA_
not modelled
12.7
16
PDB header: signaling proteinChain: A: PDB Molecule: nitrogen assimilation regulatory protein;PDBTitle: crystal structure of ntrx from brucella abortus in complex with atp2 processed with the crystaldirect automated mounting and cryo-cooling3 technology
68 c3u0oA_
not modelled
12.5
20
PDB header: transferaseChain: A: PDB Molecule: selenide, water dikinase;PDBTitle: the crystal structure of selenophosphate synthetase from e. coli
69 c3crnA_
not modelled
12.2
12
PDB header: signaling proteinChain: A: PDB Molecule: response regulator receiver domain protein, chey-like;PDBTitle: crystal structure of response regulator receiver domain protein (chey-2 like) from methanospirillum hungatei jf-1
70 d1clia1
not modelled
12.1
12
Fold: Bacillus chorismate mutase-likeSuperfamily: PurM N-terminal domain-likeFamily: PurM N-terminal domain-like
71 c4s05B_
not modelled
12.0
20
PDB header: transcription/dnaChain: B: PDB Molecule: dna-binding transcriptional regulator basr;PDBTitle: crystal structure of klebsiella pneumoniae pmra in complex with pmra2 box dna
72 d2zoda1
not modelled
11.9
19
Fold: Bacillus chorismate mutase-likeSuperfamily: PurM N-terminal domain-likeFamily: PurM N-terminal domain-like
73 d2g50a3
not modelled
11.8
19
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: PK C-terminal domain-likeFamily: Pyruvate kinase, C-terminal domain
74 d1dbwa_
not modelled
11.7
14
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related
75 c3x43F_
not modelled
11.6
10
PDB header: transferaseChain: F: PDB Molecule: o-ureido-l-serine synthase;PDBTitle: crystal structure of o-ureido-l-serine synthase
76 d2zdra2
not modelled
11.5
19
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: NeuB-like
77 c3w9sB_
not modelled
11.5
18
PDB header: signaling protein/antimicrobial proteinChain: B: PDB Molecule: ompr family response regulator in two-component regulatoryPDBTitle: crystal structure analysis of the n-terminal receiver domain of2 response regulator pmra
78 c1b74A_
not modelled
11.4
8
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: glutamate racemase from aquifex pyrophilus
79 c3cz5B_
not modelled
11.4
18
PDB header: transcription regulatorChain: B: PDB Molecule: two-component response regulator, luxr family;PDBTitle: crystal structure of two-component response regulator, luxr family,2 from aurantimonas sp. si85-9a1
80 c4e6fB_
not modelled
11.4
19
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a duf4468 family protein (bacova_04320) from2 bacteroides ovatus atcc 8483 at 1.49 a resolution
81 c5mg5P_
not modelled
11.1
15
PDB header: transferaseChain: P: PDB Molecule: hydroxymethylglutaryl-coa synthase;PDBTitle: a multi-component acyltransferase phlabc from pseudomonas protegens2 soaked with the monoacetylphloroglucinol (mapg)
82 d2e7ya1
not modelled
11.1
4
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: RNase Z-like
83 c5ijwA_
not modelled
11.1
11
PDB header: isomeraseChain: A: PDB Molecule: glutamate racemase;PDBTitle: glutamate racemase (muri) from mycobacterium smegmatis with bound d-2 glutamate, 1.8 angstrom resolution, x-ray diffraction
84 c3jteA_
not modelled
10.8
13
PDB header: protein bindingChain: A: PDB Molecule: response regulator receiver protein;PDBTitle: crystal structure of response regulator receiver domain protein from2 clostridium thermocellum
85 d1pkla3
not modelled
10.6
12
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: PK C-terminal domain-likeFamily: Pyruvate kinase, C-terminal domain
86 d1xpma1
not modelled
10.6
13
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like
87 c1w5fA_
not modelled
10.5
17
PDB header: cell divisionChain: A: PDB Molecule: cell division protein ftsz;PDBTitle: ftsz, t7 mutated, domain swapped (t. maritima)
88 c2xdqB_
not modelled
10.4
17
PDB header: oxidoreductaseChain: B: PDB Molecule: light-independent protochlorophyllide reductase subunit b;PDBTitle: dark operative protochlorophyllide oxidoreductase (chln-chlb)2 complex
89 c3aerC_
not modelled
10.3
17
PDB header: oxidoreductaseChain: C: PDB Molecule: light-independent protochlorophyllide reductase subunit n;PDBTitle: structure of the light-independent protochlorophyllide reductase2 catalyzing a key reduction for greening in the dark
90 c2yihA_
not modelled
10.3
33
PDB header: hydrolaseChain: A: PDB Molecule: cel44c;PDBTitle: structure of a paenibacillus polymyxa xyloglucanase from gh2 family 44 with xyloglucan
91 d1liua3
not modelled
10.2
17
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: PK C-terminal domain-likeFamily: Pyruvate kinase, C-terminal domain
92 c6dzkY_
not modelled
10.2
21
PDB header: ribosomeChain: Y: PDB Molecule: ribosome hibernation promoting factor;PDBTitle: cryo-em structure of mycobacterium smegmatis c(minus) 30s ribosomal2 subunit with mpy
93 c6grgD_
not modelled
9.8
15
PDB header: biosynthetic proteinChain: D: PDB Molecule: microcin b17-processing protein mcbd;PDBTitle: e. coli microcin synthetase mcbbcd complex with pro-mccb17, adp and2 phosphate bound
94 c1ys7B_
not modelled
9.5
20
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulatory protein prra;PDBTitle: crystal structure of the response regulator protein prra complexed2 with mg2+
95 d2uube2
not modelled
9.4
37
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Ribosomal S5 protein, N-terminal domain
96 c4fylB_
not modelled
9.4
8
PDB header: translationChain: B: PDB Molecule: ribosome hibernation protein yhbh;PDBTitle: high-resolution x-ray structure of hpf from vibrio cholerae
97 d1yb2a1
not modelled
9.1
20
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: tRNA(1-methyladenosine) methyltransferase-like
98 c1yb2A_
not modelled
9.1
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ta0852;PDBTitle: structure of a putative methyltransferase from thermoplasma2 acidophilum.
99 c5ohxB_
not modelled
9.0
11
PDB header: lyaseChain: B: PDB Molecule: cystathionine beta-synthase;PDBTitle: structure of active cystathionine b-synthase from apis mellifera