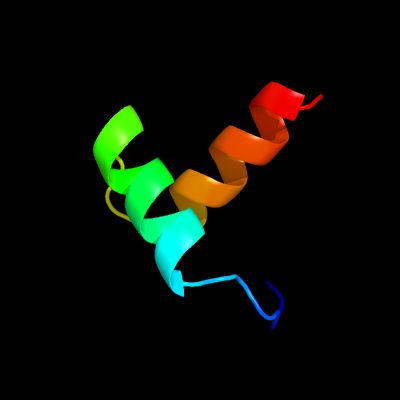
1 c6a7vU_
97.7
46
PDB header: toxin/antitoxinChain: U: PDB Molecule: antitoxin vapb11;PDBTitle: crystal structure of mycobacterium tuberculosis vapbc11 toxin-2 antitoxin complex
2 c2vdwA_
63.1
29
PDB header: transferaseChain: A: PDB Molecule: vaccinia virus capping enzyme d1 subunit;PDBTitle: guanosine n7 methyl-transferase sub-complex (d1-d12) of the2 vaccinia virus mrna capping enzyme
3 c6q56C_
30.6
29
PDB header: rna binding proteinChain: C: PDB Molecule: trna (adenine(22)-n(1))-methyltransferase;PDBTitle: crystal structure of the b. subtilis m1a22 trna methyltransferase trmk
4 c2mxdA_
25.2
64
PDB header: viral proteinChain: A: PDB Molecule: viral protein genome-linked;PDBTitle: solution structure of vpg of porcine sapovirus
5 c2m4hA_
24.8
58
PDB header: viral proteinChain: A: PDB Molecule: feline calicivirus vpg protein;PDBTitle: solution structure of the core domain (10-76) of the feline2 calicivirus vpg protein
6 c3fuxB_
24.0
21
PDB header: transferaseChain: B: PDB Molecule: dimethyladenosine transferase;PDBTitle: t. thermophilus 16s rrna a1518 and a1519 methyltransferase (ksga) in2 complex with 5'-methylthioadenosine in space group p212121
7 d2hzaa1
18.7
36
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like
8 d2f8la1
18.3
26
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: N-6 DNA Methylase-like
9 c4ponB_
17.0
17
PDB header: transferaseChain: B: PDB Molecule: putative rna methylase;PDBTitle: the crystal structure of a putative sam-dependent methyltransferase,2 ytqb, from bacillus subtilis
10 d1o54a_
16.9
23
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: tRNA(1-methyladenosine) methyltransferase-like
11 c1dl5A_
16.7
26
PDB header: transferaseChain: A: PDB Molecule: protein-l-isoaspartate o-methyltransferase;PDBTitle: protein-l-isoaspartate o-methyltransferase
12 c3g07C_
15.6
22
PDB header: transferaseChain: C: PDB Molecule: 7sk snrna methylphosphate capping enzyme;PDBTitle: methyltransferase domain of human bicoid-interacting protein 3 homolog2 (drosophila)
13 c5oy4Y_
15.2
36
PDB header: transferaseChain: Y: PDB Molecule: proto-oncogene frat1;PDBTitle: gsk3beta complex with n-(6-(3,4-dihydroxyphenyl)-1h-pyrazolo[3,4-2 b]pyridin-3-yl)acetamide
14 c3zrkX_
14.5
36
PDB header: transferase/peptideChain: X: PDB Molecule: proto-oncogene frat1;PDBTitle: identification of 2-(4-pyridyl)thienopyridinones as gsk-3beta2 inhibitors
15 c3zrlX_
14.3
36
PDB header: transferase/peptideChain: X: PDB Molecule: proto-oncogene frat1;PDBTitle: identification of 2-(4-pyridyl)thienopyridinones as gsk-3beta2 inhibitors
16 c3zrmX_
14.3
36
PDB header: transferase/peptideChain: X: PDB Molecule: proto-oncogene frat1;PDBTitle: identification of 2-(4-pyridyl)thienopyridinones as gsk-2 3beta inhibitors
17 c1gngX_
13.9
36
PDB header: transferaseChain: X: PDB Molecule: frattide;PDBTitle: glycogen synthase kinase-3 beta (gsk3) complex with frattide2 peptide
18 c3zrkY_
13.4
36
PDB header: transferase/peptideChain: Y: PDB Molecule: proto-oncogene frat1;PDBTitle: identification of 2-(4-pyridyl)thienopyridinones as gsk-3beta2 inhibitors
19 c3zrlY_
13.3
36
PDB header: transferase/peptideChain: Y: PDB Molecule: proto-oncogene frat1;PDBTitle: identification of 2-(4-pyridyl)thienopyridinones as gsk-3beta2 inhibitors
20 c3zrmY_
13.3
36
PDB header: transferase/peptideChain: Y: PDB Molecule: proto-oncogene frat1;PDBTitle: identification of 2-(4-pyridyl)thienopyridinones as gsk-2 3beta inhibitors
21 c1gngY_
not modelled
13.2
36
PDB header: transferaseChain: Y: PDB Molecule: frattide;PDBTitle: glycogen synthase kinase-3 beta (gsk3) complex with frattide2 peptide
22 c4afjY_
not modelled
13.2
36
PDB header: transferase/peptideChain: Y: PDB Molecule: proto-oncogene frat1;PDBTitle: 5-aryl-4-carboxamide-1,3-oxazoles: potent and selective gsk-32 inhibitors
23 c4ckcD_
not modelled
13.1
31
PDB header: transferase/hydrolaseChain: D: PDB Molecule: mrna-capping enzyme catalytic subunit;PDBTitle: vaccinia virus capping enzyme complexed with sah (monoclinic form)
24 c3ckkA_
not modelled
13.1
29
PDB header: transferaseChain: A: PDB Molecule: trna (guanine-n(7)-)-methyltransferase;PDBTitle: crystal structure of human methyltransferase-like protein 1
25 c5oy4X_
not modelled
13.0
36
PDB header: transferaseChain: X: PDB Molecule: proto-oncogene frat1;PDBTitle: gsk3beta complex with n-(6-(3,4-dihydroxyphenyl)-1h-pyrazolo[3,4-2 b]pyridin-3-yl)acetamide
26 c2fk8A_
not modelled
12.5
12
PDB header: transferaseChain: A: PDB Molecule: methoxy mycolic acid synthase 4;PDBTitle: crystal structure of hma (mmaa4) from mycobacterium tuberculosis2 complexed with s-adenosylmethionine
27 c2kz3A_
not modelled
12.4
34
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein rad51l3;PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for human rad51d2 from 1 to 83
28 c4afjX_
not modelled
12.1
36
PDB header: transferase/peptideChain: X: PDB Molecule: proto-oncogene frat1;PDBTitle: 5-aryl-4-carboxamide-1,3-oxazoles: potent and selective gsk-32 inhibitors
29 d1qama_
not modelled
11.4
21
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: rRNA adenine dimethylase-like
30 c1x37A_
not modelled
11.4
37
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent protease la 1;PDBTitle: structure of bacillus subtilis lon protease ssd domain
31 c3bgvC_
not modelled
10.2
35
PDB header: transferaseChain: C: PDB Molecule: mrna cap guanine-n7 methyltransferase;PDBTitle: crystal structure of mrna cap guanine-n7 methyltransferase2 in complex with sah
32 d1d2ha_
not modelled
10.1
13
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Glycine N-methyltransferase
33 c6ifsB_
not modelled
9.8
23
PDB header: transferaseChain: B: PDB Molecule: ribosomal rna small subunit methyltransferase a;PDBTitle: ksga from bacillus subtilis 168
34 d2fcaa1
not modelled
9.5
21
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: TrmB-like
35 d1knza_
not modelled
9.3
35
Fold: NSP3 homodimerSuperfamily: NSP3 homodimerFamily: NSP3 homodimer
36 c2h1rA_
not modelled
8.3
29
PDB header: transferaseChain: A: PDB Molecule: dimethyladenosine transferase, putative;PDBTitle: crystal structure of a dimethyladenosine transferase from2 plasmodium falciparum
37 d1m6ya2
not modelled
8.1
22
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: MraW-like putative methyltransferases
38 c1xrxD_
not modelled
7.7
67
PDB header: replication inhibitorChain: D: PDB Molecule: seqa protein;PDBTitle: crystal structure of a dna-binding protein
39 d1xrxa1
not modelled
7.7
67
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: SeqA N-terminal domain-like
40 d2nn6h3
not modelled
7.6
38
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)
41 c1m6yA_
not modelled
7.6
22
PDB header: transferaseChain: A: PDB Molecule: s-adenosyl-methyltransferase mraw;PDBTitle: crystal structure analysis of tm0872, a putative sam-dependent2 methyltransferase, complexed with sah
42 c3uzuA_
not modelled
7.5
30
PDB header: transferaseChain: A: PDB Molecule: ribosomal rna small subunit methyltransferase a;PDBTitle: the structure of the ribosomal rna small subunit methyltransferase a2 from burkholderia pseudomallei
43 c3grrA_
not modelled
7.5
23
PDB header: transferaseChain: A: PDB Molecule: dimethyladenosine transferase;PDBTitle: crystal structure of the complex between s-adenosyl homocysteine and2 methanocaldococcus jannaschi dim1.
44 c3fydA_
not modelled
7.5
23
PDB header: transferaseChain: A: PDB Molecule: probable dimethyladenosine transferase;PDBTitle: crystal structure of dim1 from the thermophilic archeon,2 methanocaldococcus jannaschi
45 c2kxwB_
not modelled
7.3
44
PDB header: calcium-binding protein/metal transportChain: B: PDB Molecule: sodium channel protein type 2 subunit alpha;PDBTitle: structure of the c-domain fragment of apo calmodulin bound to the iq2 motif of nav1.2
46 c2m5eB_
not modelled
7.3
44
PDB header: calcium-binding protein/metal transportChain: B: PDB Molecule: sodium channel protein type 2 subunit alpha;PDBTitle: structure of the c-domain of calcium-saturated calmodulin bound to the2 iq motif of nav1.2
47 c4jxjA_
not modelled
7.2
29
PDB header: transferaseChain: A: PDB Molecule: ribosomal rna small subunit methyltransferase a;PDBTitle: crystal structure of ribosomal rna small subunit methyltransferase a2 from rickettsia bellii determined by iodide sad phasing
48 c4aoyD_
not modelled
7.2
28
PDB header: oxidoreductaseChain: D: PDB Molecule: isocitrate dehydrogenase [nadp];PDBTitle: open ctidh. the complex structures of isocitrate dehydrogenase from2 clostridium thermocellum and desulfotalea psychrophila, support a new3 active site locking mechanism
49 c6butB_
not modelled
6.9
45
PDB header: membrane proteinChain: B: PDB Molecule: sodium channel protein type 2 subunit alpha;PDBTitle: solution structure of full-length apo mammalian calmodulin bound to2 the iq motif of the human voltage-gated sodium channel nav1.2
50 d2ieaa3
not modelled
6.9
19
Fold: TK C-terminal domain-likeSuperfamily: TK C-terminal domain-likeFamily: Transketolase C-terminal domain-like
51 c3mznA_
not modelled
6.9
27
PDB header: lyaseChain: A: PDB Molecule: glucarate dehydratase;PDBTitle: crystal structure of probable glucarate dehydratase from2 chromohalobacter salexigens dsm 3043
52 c1q5vB_
not modelled
6.8
36
PDB header: transcriptionChain: B: PDB Molecule: nickel responsive regulator;PDBTitle: apo-nikr
53 d2ahme1
not modelled
6.8
20
Fold: Coronavirus NSP8-likeSuperfamily: Coronavirus NSP8-likeFamily: Coronavirus NSP8-like
54 d2fk8a1
not modelled
6.5
15
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Mycolic acid cyclopropane synthase
55 c3vjzB_
not modelled
6.5
21
PDB header: gene regulationChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of the dna mimic protein dmp19
56 c2pbfA_
not modelled
6.3
21
PDB header: transferaseChain: A: PDB Molecule: protein-l-isoaspartate o-methyltransferase beta-aspartatePDBTitle: crystal structure of a putative protein-l-isoaspartate o-2 methyltransferase beta-aspartate methyltransferase (pcmt) from3 plasmodium falciparum in complex with s-adenosyl-l-homocysteine
57 c1mjeB_
not modelled
6.2
46
PDB header: gene regulation/antitumor protein/dnaChain: B: PDB Molecule: deleted in split hand/split foot protein 1;PDBTitle: structure of a brca2-dss1-ssdna complex
58 c2ahmG_
not modelled
6.2
20
PDB header: viral protein, replicationChain: G: PDB Molecule: replicase polyprotein 1ab, heavy chain;PDBTitle: crystal structure of sars-cov super complex of non-structural2 proteins: the hexadecamer
59 c4gitA_
not modelled
6.2
35
PDB header: hydrolaseChain: A: PDB Molecule: lon protease;PDBTitle: crystal structure of alpha sub-domain of lon protease from2 brevibacillus thermoruber
60 d1yzha1
not modelled
6.2
15
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: TrmB-like
61 c4dckA_
not modelled
6.2
36
PDB header: transport protein/signaling proteinChain: A: PDB Molecule: sodium channel protein type 5 subunit alpha;PDBTitle: crystal structure of the c-terminus of voltage-gated sodium channel in2 complex with fgf13 and cam
62 c4rwzA_
not modelled
6.1
19
PDB header: transferaseChain: A: PDB Molecule: putative rrna methyltransferase;PDBTitle: crystal structure of the antibiotic-resistance methyltransferase kmr
63 c2p7iB_
not modelled
6.1
13
PDB header: transferaseChain: B: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a sam dependent methyl-transferase type 12 family2 protein (eca1738) from pectobacterium atrosepticum scri1043 at 1.74 a3 resolution
64 c5gjqY_
not modelled
6.1
54
PDB header: hydrolaseChain: Y: PDB Molecule: 26s proteasome complex subunit dss1;PDBTitle: structure of the human 26s proteasome bound to usp14-ubal
65 d2ifta1
not modelled
6.0
18
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: YhhF-like
66 c3g5tA_
not modelled
5.9
12
PDB header: transferaseChain: A: PDB Molecule: trans-aconitate 3-methyltransferase;PDBTitle: crystal structure of trans-aconitate 3-methyltransferase from yeast
67 c3p2kA_
not modelled
5.8
11
PDB header: transferaseChain: A: PDB Molecule: 16s rrna methylase;PDBTitle: structure of an antibiotic related methyltransferase
68 c5do0A_
not modelled
5.8
24
PDB header: transferaseChain: A: PDB Molecule: protein lysine methyltransferase 1;PDBTitle: the structure of pkmt1 from rickettsia prowazekii
69 c2w6hG_
not modelled
5.7
37
PDB header: hydrolaseChain: G: PDB Molecule: atp synthase subunit gamma, mitochondrial;PDBTitle: low resolution structures of bovine mitochondrial f1-atpase during2 controlled dehydration: hydration state 4a.
70 c2ozvA_
not modelled
5.7
15
PDB header: transferaseChain: A: PDB Molecule: hypothetical protein atu0636;PDBTitle: crystal structure of a predicted o-methyltransferase, protein atu6362 from agrobacterium tumefaciens.
71 c1yb2A_
not modelled
5.7
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ta0852;PDBTitle: structure of a putative methyltransferase from thermoplasma2 acidophilum.
72 d1yb2a1
not modelled
5.7
21
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: tRNA(1-methyladenosine) methyltransferase-like
73 c3d2lC_
not modelled
5.6
11
PDB header: transferaseChain: C: PDB Molecule: sam-dependent methyltransferase;PDBTitle: crystal structure of sam-dependent methyltransferase (zp_00538691.1)2 from exiguobacterium sp. 255-15 at 1.90 a resolution
74 c3sm3A_
not modelled
5.6
17
PDB header: transferaseChain: A: PDB Molecule: sam-dependent methyltransferases;PDBTitle: crystal structure of sam-dependent methyltransferases q8puk2_metma2 from methanosarcina mazei. northeast structural genomics consortium3 target mar262.
75 c6djyD_
not modelled
5.5
25
PDB header: virusChain: D: PDB Molecule: turret protein;PDBTitle: fako virus
76 c6daeC_
not modelled
5.5
40
PDB header: calcium binding protein/membrane proteinChain: C: PDB Molecule: voltage-dependent l-type calcium channel subunit alpha-1c;PDBTitle: 2.0 angstrom crystal structure of the d95v ca/cam:cav1.2 iq domain2 complex
77 d1qyra_
not modelled
5.5
23
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: rRNA adenine dimethylase-like
78 c2ns5A_
not modelled
5.4
55
PDB header: signaling proteinChain: A: PDB Molecule: partitioning-defective 3 homolog;PDBTitle: the conserved n-terminal domain of par-3 adopts a novel pb1-2 like structure required for par-3 oligomerization and3 apical membrane localization
79 c2m4gA_
not modelled
5.3
86
PDB header: viral proteinChain: A: PDB Molecule: murine norovirus vpg protein;PDBTitle: solution structure of the core domain (11-85) of the murine norovirus2 vpg protein
80 c2bj3D_
not modelled
5.3
19
PDB header: transcriptionChain: D: PDB Molecule: nickel responsive regulator;PDBTitle: nikr-apo
81 c3gdhC_
not modelled
5.2
29
PDB header: transferaseChain: C: PDB Molecule: trimethylguanosine synthase homolog;PDBTitle: methyltransferase domain of human trimethylguanosine synthase 1 (tgs1)2 bound to m7gtp and adenosyl-homocysteine (active form)
82 c3fmtF_
not modelled
5.2
67
PDB header: replication inhibitor/dnaChain: F: PDB Molecule: protein seqa;PDBTitle: crystal structure of seqa bound to dna
83 c4fsxB_
not modelled
5.1
19
PDB header: transferaseChain: B: PDB Molecule: dna (cytosine-5)-methyltransferase 1;PDBTitle: crystal structure of se-substituted zea mays zmet2 in complex with sah
84 c2be6F_
not modelled
5.1
40
PDB header: membrane proteinChain: F: PDB Molecule: voltage-dependent l-type calcium channel alpha-1c subunit;PDBTitle: 2.0 a crystal structure of the cav1.2 iq domain-ca/cam complex