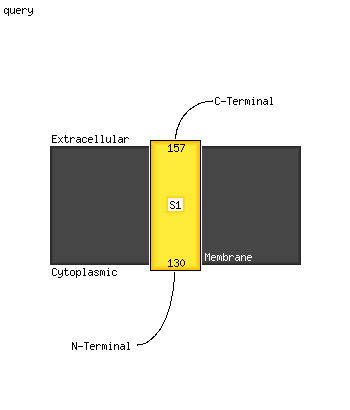
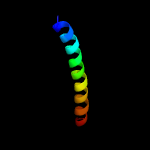
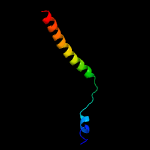
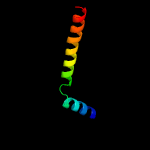
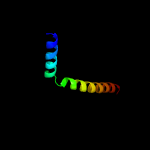
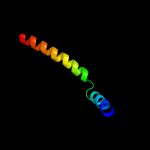
1 c5ch4E_
99.8
26
PDB header: protein transportChain: E: PDB Molecule: protein translocase subunit sece;PDBTitle: peptide-bound state of thermus thermophilus secyeg
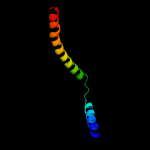
2 c5eulE_
99.8
49
PDB header: protein transportChain: E: PDB Molecule: preprotein translocase sece subunit;PDBTitle: structure of the seca-secy complex with a translocating polypeptide2 substrate
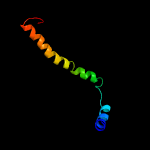
3 c2akhZ_
99.8
32
PDB header: protein transportChain: Z: PDB Molecule: preprotein translocase sece subunit;PDBTitle: normal mode-based flexible fitted coordinates of a non-translocating2 secyeg protein-conducting channel into the cryo-em map of a secyeg-3 nascent chain-70s ribosome complex from e. coli
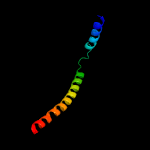
4 c3dinD_
99.7
35
PDB header: membrane protein, protein transportChain: D: PDB Molecule: preprotein translocase subunit sece;PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
5 c2zjsE_
99.0
28
PDB header: protein transport/immune systemChain: E: PDB Molecule: preprotein translocase sece subunit;PDBTitle: crystal structure of secye translocon from thermus thermophilus with a2 fab fragment
6 c2zqpE_
98.9
27
PDB header: protein transportChain: E: PDB Molecule: preprotein translocase sece subunit;PDBTitle: crystal structure of secye translocon from thermus2 thermophilus
7 c3dl8D_
97.9
21
PDB header: protein transportChain: D: PDB Molecule: sece;PDBTitle: structure of the complex of aquifex aeolicus secyeg and bacillus2 subtilis seca
8 c3dl8C_
97.9
21
PDB header: protein transportChain: C: PDB Molecule: sece;PDBTitle: structure of the complex of aquifex aeolicus secyeg and bacillus2 subtilis seca
9 d1rhzb_
94.9
9
Fold: Single transmembrane helixSuperfamily: Preprotein translocase SecE subunitFamily: Preprotein translocase SecE subunit
10 c2wwbB_
93.7
14
PDB header: ribosomeChain: B: PDB Molecule: protein transport protein sec61 subunit gamma;PDBTitle: cryo-em structure of the mammalian sec61 complex bound to the actively2 translating wheat germ 80s ribosome
11 c2ww9B_
92.1
9
PDB header: ribosomeChain: B: PDB Molecule: protein transport protein sss1;PDBTitle: cryo-em structure of the active yeast ssh1 complex bound to the yeast2 80s ribosome
12 d1rh5b_
80.1
8
Fold: Single transmembrane helixSuperfamily: Preprotein translocase SecE subunitFamily: Preprotein translocase SecE subunit
13 c4cdiC_
75.8
17
PDB header: membrane proteinChain: C: PDB Molecule: predicted protein;PDBTitle: crystal structure of acrb-acrz complex
14 d1wh8a_
62.8
21
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: CUT domain
15 c1zrtD_
60.8
12
PDB header: oxidoreductase/metal transportChain: D: PDB Molecule: cytochrome c1;PDBTitle: rhodobacter capsulatus cytochrome bc1 complex with2 stigmatellin bound
16 c3mp7B_
57.9
13
PDB header: protein transportChain: B: PDB Molecule: preprotein translocase subunit sece;PDBTitle: lateral opening of a translocon upon entry of protein suggests the2 mechanism of insertion into membranes
17 c2fynH_
57.6
14
PDB header: oxidoreductaseChain: H: PDB Molecule: cytochrome c1;PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
18 c1p84D_
52.9
11
PDB header: oxidoreductaseChain: D: PDB Molecule: cytochrome c1, heme protein;PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
19 c3cwbQ_
47.7
19
PDB header: oxidoreductaseChain: Q: PDB Molecule: mitochondrial cytochrome c1, heme protein;PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
20 c2yiuE_
45.7
19
PDB header: oxidoreductaseChain: E: PDB Molecule: cytochrome c1, heme protein;PDBTitle: x-ray structure of the dimeric cytochrome bc1 complex from2 the soil bacterium paracoccus denitrificans at 2.73 angstrom resolution
21 c1qcrD_
not modelled
39.4
17
PDB header: oxidoreductaseChain: D: PDB Molecule: ubiquinol cytochrome c oxidoreductase;PDBTitle: crystal structure of bovine mitochondrial cytochrome bc12 complex, alpha carbon atoms only
22 c2kogA_
not modelled
32.5
18
PDB header: membrane proteinChain: A: PDB Molecule: vesicle-associated membrane protein 2;PDBTitle: lipid-bound synaptobrevin solution nmr structure
23 c2eqzA_
not modelled
24.2
17
PDB header: transcriptionChain: A: PDB Molecule: high mobility group protein b3;PDBTitle: solution structure of the first hmg-box domain from high2 mobility group protein b3
24 c3qf4B_
not modelled
22.3
15
PDB header: transport proteinChain: B: PDB Molecule: uncharacterized abc transporter atp-binding proteinPDBTitle: crystal structure of a heterodimeric abc transporter in its inward-2 facing conformation
25 d2e74b1
not modelled
21.4
19
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
26 c4h1wB_
not modelled
20.9
24
PDB header: hydrolase/hydrolase regulatorChain: B: PDB Molecule: sarcolipin;PDBTitle: e1 structure of the (sr) ca2+-atpase in complex with sarcolipin
27 c3w5aC_
not modelled
20.9
24
PDB header: metal transport/membrane proteinChain: C: PDB Molecule: sarcolipin;PDBTitle: crystal structure of the calcium pump and sarcolipin from rabbit fast2 twitch skeletal muscle in the e1.mg2+ state
28 c6fs4A_
not modelled
20.0
31
PDB header: antimicrobial proteinChain: A: PDB Molecule: alpha-s2-casein;PDBTitle: nmr structure of casocidin-ii antimicrobial peptide in 60% tfe
29 d2h8pc1
not modelled
19.6
15
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels
30 c3hd7A_
not modelled
19.5
8
PDB header: exocytosisChain: A: PDB Molecule: vesicle-associated membrane protein 2;PDBTitle: helical extension of the neuronal snare complex into the membrane,2 spacegroup c 1 2 1
31 d1v54d_
not modelled
19.0
19
Fold: Single transmembrane helixSuperfamily: Mitochondrial cytochrome c oxidase subunit IVFamily: Mitochondrial cytochrome c oxidase subunit IV
32 c1jdmA_
not modelled
17.4
24
PDB header: membrane proteinChain: A: PDB Molecule: sarcolipin;PDBTitle: nmr structure of sarcolipin
33 c4gn0D_
not modelled
17.3
13
PDB header: signaling proteinChain: D: PDB Molecule: hamp domain of af1503;PDBTitle: de novo phasing of a hamp-complex using an improved arcimboldo method
34 d2axth1
not modelled
16.7
20
Fold: Single transmembrane helixSuperfamily: Photosystem II 10 kDa phosphoprotein PsbHFamily: PsbH-like
35 c2axtH_
not modelled
16.7
20
PDB header: electron transportChain: H: PDB Molecule: photosystem ii reaction center h protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
36 c5gasN_
not modelled
16.3
18
PDB header: hydrolaseChain: N: PDB Molecule: archaeal/vacuolar-type h+-atpase subunit i;PDBTitle: thermus thermophilus v/a-atpase, conformation 2
37 c2wg7B_
not modelled
16.3
13
PDB header: hydrolaseChain: B: PDB Molecule: putative phospholipase a2;PDBTitle: structure of oryza sativa (rice) pla2
38 c2wwbC_
not modelled
16.0
25
PDB header: ribosomeChain: C: PDB Molecule: protein transport protein sec61 subunit beta;PDBTitle: cryo-em structure of the mammalian sec61 complex bound to the actively2 translating wheat germ 80s ribosome
39 c2ehbD_
not modelled
15.9
18
PDB header: signalling protein/transferaseChain: D: PDB Molecule: cbl-interacting serine/threonine-protein kinase 24;PDBTitle: the structure of the c-terminal domain of the protein kinase atsos22 bound to the calcium sensor atsos3
40 d1j3xa_
not modelled
15.8
14
Fold: HMG-boxSuperfamily: HMG-boxFamily: HMG-box
41 c1j3xA_
not modelled
15.8
14
PDB header: dna binding proteinChain: A: PDB Molecule: high mobility group protein 2;PDBTitle: solution structure of the n-terminal domain of the hmgb2
42 c2co9A_
not modelled
15.5
16
PDB header: transcriptionChain: A: PDB Molecule: thymus high mobility group box protein tox;PDBTitle: solution structure of the hmg_box domain of thymus high2 mobility group box protein tox from mouse
43 c6gy4B_
not modelled
15.4
18
PDB header: rna binding proteinChain: B: PDB Molecule: protein bicaudal c homolog 1;PDBTitle: crystal structure of the n-terminal kh domain of human bicc1
44 c3fwbB_
not modelled
15.3
17
PDB header: cell cycle, transcriptionChain: B: PDB Molecule: nuclear mrna export protein sac3;PDBTitle: sac3:sus1:cdc31 complex
45 c4il3B_
not modelled
14.9
8
PDB header: hydrolaseChain: B: PDB Molecule: ste24p;PDBTitle: crystal structure of s. mikatae ste24p
46 c2e6oA_
not modelled
14.8
13
PDB header: transcription, cell cycleChain: A: PDB Molecule: hmg box-containing protein 1;PDBTitle: solution structure of the hmg box domain from human hmg-box2 transcription factor 1
47 d1wh6a_
not modelled
14.2
23
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: CUT domain
48 c2y69Q_
not modelled
13.6
19
PDB header: electron transportChain: Q: PDB Molecule: cytochrome c oxidase subunit 4 isoform 1;PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular oxygen
49 c6o7ua_
not modelled
13.6
10
PDB header: membrane proteinChain: A: PDB Molecule: PDBTitle: saccharomyces cerevisiae v-atpase stv1-vo
50 c4hd1A_
not modelled
13.2
13
PDB header: transferaseChain: A: PDB Molecule: squalene synthase hpnc;PDBTitle: crystal structure of squalene synthase hpnc from alicyclobacillus2 acidocaldarius
51 c2ndjA_
not modelled
13.0
19
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily e member 3;PDBTitle: structural basis for kcne3 and estrogen modulation of the kcnq12 channel
52 c2m0qA_
not modelled
11.7
16
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily e member 2;PDBTitle: solution nmr analysis of intact kcne2 in detergent micelles2 demonstrate a straight transmembrane helix
53 c1yq3C_
not modelled
11.5
15
PDB header: oxidoreductaseChain: C: PDB Molecule: succinate dehydrogenase cytochrome b, large subunit;PDBTitle: avian respiratory complex ii with oxaloacetate and ubiquinone
54 c2m3aA_
not modelled
9.9
22
PDB header: dna binding proteinChain: A: PDB Molecule: protein knl-2;PDBTitle: nmr solution structure of a myb-like dna binding domain of knl-2 from2 c. elegans
55 d2ezwa1
not modelled
9.5
32
Fold: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunitSuperfamily: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunitFamily: Dimerization-anchoring domain of cAMP-dependent PK regulatory subunit
56 c2n28A_
not modelled
9.0
18
PDB header: viral proteinChain: A: PDB Molecule: protein vpu;PDBTitle: solid-state nmr structure of vpu
57 c2gofA_
not modelled
8.7
18
PDB header: viral proteinChain: A: PDB Molecule: vpu protein;PDBTitle: three-dimensional structure of the trans-membrane domain of2 vpu from hiv-1 in aligned phospholipid bicelles
58 c2gohA_
not modelled
8.7
18
PDB header: viral proteinChain: A: PDB Molecule: vpu protein;PDBTitle: three-dimensional structure of the trans-membrane domain of2 vpu from hiv-1 in aligned phospholipid bicelles
59 c1pi8A_
not modelled
8.7
18
PDB header: viral proteinChain: A: PDB Molecule: vpu protein;PDBTitle: structure of the channel-forming trans-membrane domain of2 virus protein "u" (vpu) from hiv-1
60 c1pjeA_
not modelled
8.7
18
PDB header: viral proteinChain: A: PDB Molecule: vpu protein;PDBTitle: structure of the channel-forming trans-membrane domain of2 virus protein "u"(vpu) from hiv-1
61 c1pi7A_
not modelled
8.7
18
PDB header: viral proteinChain: A: PDB Molecule: vpu protein;PDBTitle: structure of the channel-forming trans-membrane domain of2 virus protein "u" (vpu) from hiv-1
62 d1q90d_
not modelled
8.6
26
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
63 c2wmhA_
not modelled
8.4
11
PDB header: hydrolaseChain: A: PDB Molecule: fucolectin-related protein;PDBTitle: crystal structure of the catalytic module of a family 982 glycoside hydrolase from streptococcus pneumoniae tigr4 in3 complex with the h-disaccharide blood group antigen.
64 c2m59B_
not modelled
8.4
12
PDB header: transferaseChain: B: PDB Molecule: vascular endothelial growth factor receptor 2;PDBTitle: spatial structure of dimeric vegfr2 membrane domain in dpc micelles
65 c2m59A_
not modelled
8.4
12
PDB header: transferaseChain: A: PDB Molecule: vascular endothelial growth factor receptor 2;PDBTitle: spatial structure of dimeric vegfr2 membrane domain in dpc micelles
66 c6m97A_
not modelled
8.3
7
PDB header: transport proteinChain: A: PDB Molecule: chimera protein of high affinity copper uptake protein 1PDBTitle: crystal structure of the high-affinity copper transporter ctr1
67 d2axti1
not modelled
7.9
24
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein I, PsbIFamily: PsbI-like
68 c3a0hi_
not modelled
7.9
24
PDB header: electron transportChain: I: PDB Molecule: photosystem ii reaction center protein i;PDBTitle: crystal structure of i-substituted photosystem ii complex
69 c4f9kA_
not modelled
7.8
14
PDB header: transferase regulatorChain: A: PDB Molecule: camp-dependent protein kinase type i-beta regulatoryPDBTitle: crystal structure of human camp-dependent protein kinase type i-beta2 regulatory subunit (fragment 11-73), northeast structural genomics3 consortium (nesg) target hr8613a
70 c3ipdB_
not modelled
7.7
9
PDB header: exocytosisChain: B: PDB Molecule: syntaxin-1a;PDBTitle: helical extension of the neuronal snare complex into the2 membrane, spacegroup i 21 21 21
71 c2ly4A_
not modelled
7.5
19
PDB header: nuclear protein/antitumour proteinChain: A: PDB Molecule: high mobility group protein b1;PDBTitle: hmgb1-facilitated p53 dna binding occurs via hmg-box/p532 transactivation domain interaction and is regulated by the acidic3 tail
72 c2wmkB_
not modelled
7.5
22
PDB header: hydrolaseChain: B: PDB Molecule: fucolectin-related protein;PDBTitle: crystal structure of the catalytic module of a family 982 glycoside hydrolase from streptococcus pneumoniae sp3-bs713 (sp3gh98) in complex with the a-lewisy pentasaccharide4 blood group antigen.
73 c4mrnB_
not modelled
7.4
17
PDB header: transport proteinChain: B: PDB Molecule: abc transporter related protein;PDBTitle: structure of a bacterial atm1-family abc transporter
74 c6fs5A_
not modelled
7.2
21
PDB header: antimicrobial proteinChain: A: PDB Molecule: alpha-s2-casein;PDBTitle: nmr structure of casocidin-i antimicrobial peptide in 60% tfe
75 c2k21A_
not modelled
7.1
29
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily ePDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
76 c3jcuI_
not modelled
6.8
18
PDB header: membrane proteinChain: I: PDB Molecule: protein photosystem ii reaction center protein i;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
77 c4y21A_
not modelled
6.7
16
PDB header: exocytosisChain: A: PDB Molecule: protein unc-13 homolog a;PDBTitle: crystal structure of munc13-1 mun domain
78 c6nd1D_
not modelled
6.6
21
PDB header: protein transportChain: D: PDB Molecule: protein transport protein sbh1;PDBTitle: cryoem structure of the sec complex from yeast
79 d1x2la1
not modelled
6.5
23
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: CUT domain
80 c2mfrA_
not modelled
6.5
22
PDB header: transferaseChain: A: PDB Molecule: insulin receptor;PDBTitle: solution structure of the transmembrane domain of the insulin receptor2 in micelles
81 c5mx2T_
not modelled
6.4
13
PDB header: oxidoreductaseChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: photosystem ii depleted of the mn4cao5 cluster at 2.55 a resolution
82 c4aw6B_
not modelled
6.4
6
PDB header: hydrolaseChain: B: PDB Molecule: caax prenyl protease 1 homolog;PDBTitle: crystal structure of the human nuclear membrane zinc metalloprotease2 zmpste24 (face1)
83 c5b66T_
not modelled
5.9
13
PDB header: electron transport, photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure analysis of photosystem ii complex
84 c5b5et_
not modelled
5.9
13
PDB header: electron transport, photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure analysis of photosystem ii complex
85 c4il6T_
not modelled
5.9
13
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: structure of sr-substituted photosystem ii
86 c5b5eT_
not modelled
5.8
13
PDB header: electron transport, photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure analysis of photosystem ii complex
87 c3wu2t_
not modelled
5.8
13
PDB header: electron transport, photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure analysis of photosystem ii complex
88 c4ub6t_
not modelled
5.8
13
PDB header: electron transport, photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: native structure of photosystem ii (dataset-1) by a femtosecond x-ray2 laser
89 c5kaft_
not modelled
5.8
13
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: rt xfel structure of photosystem ii in the dark state at 3.0 a2 resolution
90 c4ub6T_
not modelled
5.8
13
PDB header: electron transport, photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: native structure of photosystem ii (dataset-1) by a femtosecond x-ray2 laser
91 c4ub8t_
not modelled
5.8
13
PDB header: electron transport, photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: native structure of photosystem ii (dataset-2) by a femtosecond x-ray2 laser
92 c3wu2T_
not modelled
5.8
13
PDB header: electron transport, photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure analysis of photosystem ii complex
93 c5kaiT_
not modelled
5.8
13
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: nh3-bound rt xfel structure of photosystem ii 500 ms after the 2nd2 illumination (2f) at 2.8 a resolution
94 c5gtht_
not modelled
5.8
13
PDB header: photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: native xfel structure of photosystem ii (dark dataset)
95 c5v2cT_
not modelled
5.8
13
PDB header: electron transport, photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: re-refinement of crystal structure of photosystem ii complex
96 c5zznT_
not modelled
5.8
13
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of photosystem ii from an sqdg-deficient mutant of2 thermosynechococcus elongatus
97 c4ub8T_
not modelled
5.8
13
PDB header: electron transport, photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: native structure of photosystem ii (dataset-2) by a femtosecond x-ray2 laser
98 c5b66t_
not modelled
5.8
13
PDB header: electron transport, photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure analysis of photosystem ii complex
99 c5zznt_
not modelled
5.8
13
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of photosystem ii from an sqdg-deficient mutant of2 thermosynechococcus elongatus