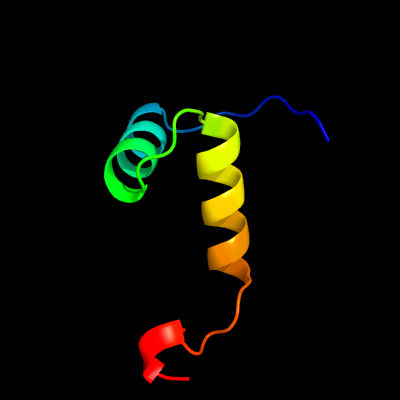
1 c6a7vU_
99.1
47
PDB header: toxin/antitoxinChain: U: PDB Molecule: antitoxin vapb11;PDBTitle: crystal structure of mycobacterium tuberculosis vapbc11 toxin-2 antitoxin complex
2 c2k5jB_
71.6
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein yiif;PDBTitle: solution structure of protein yiif from shigella flexneri2 serotype 5b (strain 8401) . northeast structural genomics3 consortium target sft1
3 c3echC_
45.8
50
PDB header: transcription, transcription regulationChain: C: PDB Molecule: 25-mer fragment of protein armr;PDBTitle: the marr-family repressor mexr in complex with its antirepressor armr
4 d1c1da2
44.8
40
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases
5 c1q5vB_
29.2
22
PDB header: transcriptionChain: B: PDB Molecule: nickel responsive regulator;PDBTitle: apo-nikr
6 c2bj3D_
27.9
19
PDB header: transcriptionChain: D: PDB Molecule: nickel responsive regulator;PDBTitle: nikr-apo
7 d1leha2
27.6
40
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases
8 c2p2uA_
23.9
9
PDB header: dna binding proteinChain: A: PDB Molecule: host-nuclease inhibitor protein gam, putative;PDBTitle: crystal structure of putative host-nuclease inhibitor protein gam from2 desulfovibrio vulgaris
9 d2bj7a1
23.7
17
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like
10 c6iyaD_
10.6
28
PDB header: antitoxinChain: D: PDB Molecule: transcriptional regulator copg family;PDBTitle: structure of the dna binding domain of antitoxin copaso
11 c1bxgA_
10.3
40
PDB header: amino acid dehydrogenaseChain: A: PDB Molecule: phenylalanine dehydrogenase;PDBTitle: phenylalanine dehydrogenase structure in ternary complex2 with nad+ and beta-phenylpropionate
12 c2ca9B_
9.9
14
PDB header: transcriptionChain: B: PDB Molecule: putative nickel-responsive regulator;PDBTitle: apo-nikr from helicobacter pylori in closed trans-2 conformation
13 c2ltuA_
9.7
23
PDB header: transferaseChain: A: PDB Molecule: 5'-amp-activated protein kinase catalytic subunit alpha-2;PDBTitle: solution structure of autoinhibitory domain of human amp-activated2 protein kinase catalytic subunit
14 c2wj9A_
8.3
31
PDB header: hydrolase inhibitorChain: A: PDB Molecule: intergenic-region protein;PDBTitle: ardb
15 c3sztB_
7.5
27
PDB header: transcriptionChain: B: PDB Molecule: quorum-sensing control repressor;PDBTitle: quorum sensing control repressor, qscr, bound to n-3-oxo-dodecanoyl-l-2 homoserine lactone
16 d2hzaa1
7.4
16
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like
17 c1lehB_
7.2
40
PDB header: oxidoreductaseChain: B: PDB Molecule: leucine dehydrogenase;PDBTitle: leucine dehydrogenase from bacillus sphaericus
18 c5zyxA_
7.0
40
PDB header: antimicrobial proteinChain: A: PDB Molecule: arg-trp-lys-arg-his-ile-ser-glu-gln-leu-arg-arg-arg-asp-PDBTitle: solution nmr structure of k30 peptide in 10 mm dioctanoyl2 phosphatidylglycerol (d8pg)
19 d1kwia_
6.7
36
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: Cathelicidin motif
20 c2klzA_
6.5
21
PDB header: hydrolaseChain: A: PDB Molecule: ataxin-3;PDBTitle: solution structure of the tandem uim domain of ataxin-3 complexed with2 ubiquitin
21 c2hv8D_
not modelled
6.4
38
PDB header: protein transportChain: D: PDB Molecule: rab11 family-interacting protein 3;PDBTitle: crystal structure of gtp-bound rab11 in complex with fip3
22 c2kmgA_
not modelled
6.2
19
PDB header: gene regulationChain: A: PDB Molecule: klca;PDBTitle: the structure of the klca and ardb proteins show a novel2 fold and antirestriction activity against type i dna3 restriction systems in vivo but not in vitro
23 c2khsB_
not modelled
6.1
23
PDB header: hydrolaseChain: B: PDB Molecule: nuclease;PDBTitle: solution structure of snase121:snase(111-143) complex
24 c4cgbA_
not modelled
6.1
33
PDB header: cell cycleChain: A: PDB Molecule: echinoderm microtubule-associated protein-like 2;PDBTitle: crystal structure of the trimerization domain of eml2
25 c2mdvB_
not modelled
6.0
42
PDB header: de novo proteinChain: B: PDB Molecule: designed protein;PDBTitle: nmr structure of beta alpha alpha 38
26 c2q2kB_
not modelled
6.0
40
PDB header: dna binding protein/dnaChain: B: PDB Molecule: hypothetical protein;PDBTitle: structure of nucleic-acid binding protein
27 c2q2kA_
not modelled
6.0
40
PDB header: dna binding protein/dnaChain: A: PDB Molecule: hypothetical protein;PDBTitle: structure of nucleic-acid binding protein
28 c4cgbE_
not modelled
6.0
63
PDB header: cell cycleChain: E: PDB Molecule: echinoderm microtubule-associated protein-like 2;PDBTitle: crystal structure of the trimerization domain of eml2
29 c2f4qA_
not modelled
5.7
15
PDB header: isomeraseChain: A: PDB Molecule: type i topoisomerase, putative;PDBTitle: crystal structure of deinococcus radiodurans topoisomerase ib
30 c6dmaD_
not modelled
5.6
33
PDB header: de novo proteinChain: D: PDB Molecule: dhd15_closed_b;PDBTitle: dhd15_closed
31 c2d7cD_
not modelled
5.6
40
PDB header: protein transportChain: D: PDB Molecule: rab11 family-interacting protein 3;PDBTitle: crystal structure of human rab11 in complex with fip3 rab-2 binding domain
32 d1vjla_
not modelled
5.3
30
Fold: Hypothetical protein TM0160Superfamily: Hypothetical protein TM0160Family: Hypothetical protein TM0160
33 d2hzab1
not modelled
5.3
19
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like
34 c4nawC_
not modelled
5.2
40
PDB header: protein transport/ligaseChain: C: PDB Molecule: autophagy-related protein 16-1;PDBTitle: crystal structure of human atg12~atg5-atg16n in complex with a2 fragment of atg3
35 c4nawG_
not modelled
5.2
40
PDB header: protein transport/ligaseChain: G: PDB Molecule: autophagy-related protein 16-1;PDBTitle: crystal structure of human atg12~atg5-atg16n in complex with a2 fragment of atg3
36 c4nawO_
not modelled
5.2
40
PDB header: protein transport/ligaseChain: O: PDB Molecule: autophagy-related protein 16-1;PDBTitle: crystal structure of human atg12~atg5-atg16n in complex with a2 fragment of atg3
37 c4gdlC_
not modelled
5.2
40
PDB header: protein bindingChain: C: PDB Molecule: autophagy-related protein 16-1;PDBTitle: crystal structure of human atg12~atg5 conjugate in complex with an n-2 terminal fragment of atg16l1
38 c4gdkC_
not modelled
5.2
40
PDB header: protein bindingChain: C: PDB Molecule: autophagy-related protein 16-1;PDBTitle: crystal structure of human atg12~atg5 conjugate in complex with an n-2 terminal fragment of atg16l1
39 c4nawK_
not modelled
5.2
40
PDB header: protein transport/ligaseChain: K: PDB Molecule: autophagy-related protein 16-1;PDBTitle: crystal structure of human atg12~atg5-atg16n in complex with a2 fragment of atg3