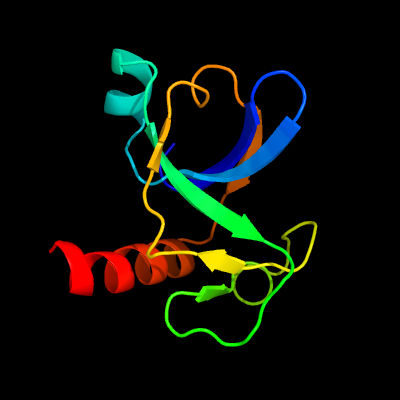
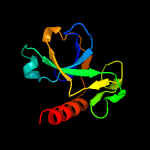
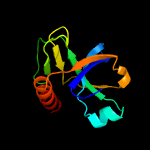
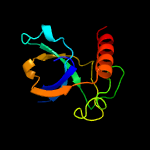
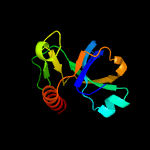
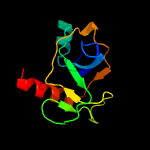
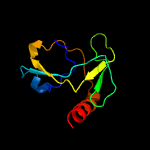
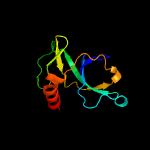
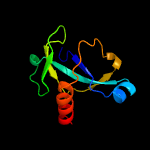
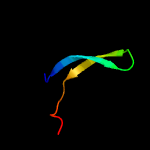
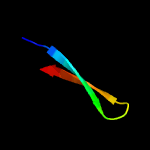
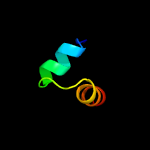
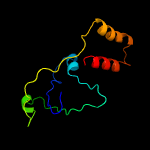
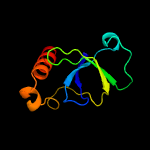
1 d1ub4a_
100.0
21
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Kid/PemK
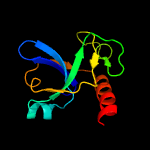
2 c5xe3B_
99.9
25
PDB header: hydrolase/antitoxinChain: B: PDB Molecule: endoribonuclease mazf4;PDBTitle: endoribonuclease in complex with its cognate antitoxin from2 mycobacterial species
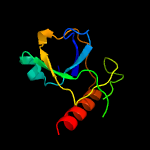
3 c4mzpC_
99.9
28
PDB header: hydrolaseChain: C: PDB Molecule: mazf mrna interferase;PDBTitle: mazf from s. aureus crystal form iii, c2221, 2.7 a
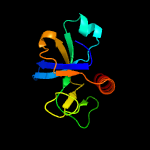
4 c5hk3B_
99.9
31
PDB header: hydrolase/dnaChain: B: PDB Molecule: endoribonuclease mazf6;PDBTitle: crystal structure of m. tuberculosis mazf-mt3 t52d-f62d mutant in2 complex with dna
5 c5hjzA_
99.9
26
PDB header: hydrolase/rnaChain: A: PDB Molecule: endoribonuclease mazf9;PDBTitle: structure of m. tuberculosis mazf-mt1 (rv2801c) in complex with rna
6 c5wygC_
99.9
27
PDB header: hydrolaseChain: C: PDB Molecule: probable endoribonuclease mazf7;PDBTitle: the crystal structure of the apo form of mtb mazf
7 d1ne8a_
99.9
28
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Kid/PemK
8 d1m1fa_
99.9
27
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Kid/PemK
9 c5ccaA_
99.9
30
PDB header: hydrolaseChain: A: PDB Molecule: endoribonuclease mazf3;PDBTitle: crystal structure of mtb toxin
10 d3vuba_
96.5
14
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: CcdB
11 c3jrzA_
95.8
12
PDB header: toxinChain: A: PDB Molecule: ccdb;PDBTitle: ccdbvfi-formii-ph5.6
12 c5ikjA_
64.8
19
PDB header: transcriptionChain: A: PDB Molecule: cryptic loci regulator 2;PDBTitle: structure of clr2 bound to the clr1 c-terminus
13 c6bwqB_
21.1
33
PDB header: metal binding proteinChain: B: PDB Molecule: pyridinium-3,5-bisthiocarboxylic acid mononucleotide nickelPDBTitle: larc2, the c-terminal domain of a cyclometallase involved in the2 synthesis of the npn cofactor of lactate racemase, in complex with3 mnctp
14 c3c19A_
20.0
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein mk0293;PDBTitle: crystal structure of protein mk0293 from methanopyrus kandleri av19
15 d1rz4a1
17.9
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Eukaryotic translation initiation factor 3 subunit 12, eIF3k, C-terminal domain
16 c3vdoB_
13.8
9
PDB header: dna binding protein/protein bindingChain: B: PDB Molecule: anti-sigma-k factor rska;PDBTitle: structure of extra-cytoplasmic function(ecf) sigma factor sigk in2 complex with its negative regulator rska from mycobacterium3 tuberculosis
17 d1okja2
12.5
50
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: YeaZ-like
18 c4p7xA_
12.4
14
PDB header: oxidoreductaseChain: A: PDB Molecule: l-proline cis-4-hydroxylase;PDBTitle: l-pipecolic acid-bound l-proline cis-4-hydroxylase
19 c3llrA_
10.8
17
PDB header: transferaseChain: A: PDB Molecule: dna (cytosine-5)-methyltransferase 3a;PDBTitle: crystal structure of the pwwp domain of human dna (cytosine-5-)-2 methyltransferase 3 alpha
20 c1cffB_
10.5
38
PDB header: calmodulinChain: B: PDB Molecule: calcium pump;PDBTitle: nmr solution structure of a complex of calmodulin with a binding2 peptide of the ca2+-pump
21 c5fmzB_
not modelled
9.8
17
PDB header: transcriptionChain: B: PDB Molecule: rna-directed rna polymerase catalytic subunit;PDBTitle: crystal structure of influenza b polymerase with bound 5' vrna
22 c4wsaB_
not modelled
9.6
17
PDB header: transferase/rnaChain: B: PDB Molecule: rna-directed rna polymerase catalytic subunit;PDBTitle: crystal structure of influenza b polymerase bound to the vrna promoter2 (flub1 form)
23 c4nohA_
not modelled
9.4
14
PDB header: lipid binding proteinChain: A: PDB Molecule: lipoprotein, putative;PDBTitle: 1.5 angstrom crystal structure of putative lipoprotein from bacillus2 anthracis.
24 c2l89A_
not modelled
9.2
17
PDB header: protein bindingChain: A: PDB Molecule: pwwp domain-containing protein 1;PDBTitle: solution structure of pdp1 pwwp domain reveals its unique binding2 sites for methylated h4k20 and dna
25 d1cxqa_
not modelled
9.1
19
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
26 c2dkkA_
not modelled
8.2
32
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome p450;PDBTitle: structure/function studies of cytochrome p450 158a1 from streptomyces2 coelicolor a3(2)
27 c2z3tD_
not modelled
8.1
9
PDB header: oxidoreductaseChain: D: PDB Molecule: cytochrome p450;PDBTitle: crystal structure of substrate free cytochrome p450 stap2 (cyp245a1)
28 c2jnsA_
not modelled
7.8
13
PDB header: unknown functionChain: A: PDB Molecule: bromodomain-containing protein 4;PDBTitle: solution structure of the bromodomain-containing protein 42 et domain
29 c4bedA_
not modelled
7.2
17
PDB header: oxygen transportChain: A: PDB Molecule: hemocyanin klh1;PDBTitle: keyhole limpet hemocyanin (klh): 9a cryoem structure and molecular2 model of the klh1 didecamer reveal the interfaces and intricate3 topology of the 160 functional units
30 d2dexx3
not modelled
6.8
15
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Peptidylarginine deiminase Pad4, catalytic C-terminal domain
31 c5cz1B_
not modelled
6.8
19
PDB header: hydrolaseChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of the catalytic core domain of mmtv integrase
32 c2jysA_
not modelled
6.7
7
PDB header: hydrolaseChain: A: PDB Molecule: protease/reverse transcriptase;PDBTitle: solution structure of simian foamy virus (mac) protease
33 d1lm8v_
not modelled
6.7
14
Fold: Prealbumin-likeSuperfamily: VHLFamily: VHL
34 c4bkwA_
not modelled
6.6
15
PDB header: receptorChain: A: PDB Molecule: zinc finger fyve domain-containing protein 9;PDBTitle: crystal structure of the c-terminal region of human zfyve9
35 d1s1fa_
not modelled
6.5
27
Fold: Cytochrome P450Superfamily: Cytochrome P450Family: Cytochrome P450
36 c2qnhv_
not modelled
6.4
47
PDB header: ribosomeChain: V: PDB Molecule: PDBTitle: interactions and dynamics of the shine-dalgarno helix in the 70s2 ribosome. this file, 2qnh, contains the 30s ribosome subunit, two3 trna, and mrna molecules. 50s ribosome subunit is in the file 1vsp.
37 c3v28U_
not modelled
6.0
47
PDB header: ribosomeChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: crystal structure of hpf bound to the 70s ribosome. this pdb entry2 contains coordinates for the 30s subunit with bound hpf of the 2nd3 ribosome in the asu
38 c4dv2U_
not modelled
5.9
47
PDB header: ribosomeChain: U: PDB Molecule: ribosomal protein thx;PDBTitle: crystal structure of the thermus thermophilus 30s ribosomal subunit2 with a 16s rrna mutation, c912a
39 c3ohdU_
not modelled
5.9
47
PDB header: ribosome/antibioticChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: structure of the thermus thermophilus ribosome complexed with2 erythromycin. this file contains the 30s subunit of one 70s ribosome.3 the entire crystal structure contains two 70s ribosomes.
40 c3ohcU_
not modelled
5.9
47
PDB header: ribosome/antibioticChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: structure of the thermus thermophilus ribosome complexed with2 erythromycin. this file contains the 30s subunit of one 70s ribosome.3 the entire crystal structure contains two 70s ribosomes.
41 c3d55A_
not modelled
5.9
7
PDB header: toxin inhibitorChain: A: PDB Molecule: uncharacterized protein rv3357/mt3465;PDBTitle: crystal structure of m. tuberculosis yefm antitoxin
42 d1uyra1
not modelled
5.8
35
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Biotin dependent carboxylase carboxyltransferase domain
43 c4dv1U_
not modelled
5.5
47
PDB header: ribosome/antibioticChain: U: PDB Molecule: ribosomal protein thx;PDBTitle: crystal structure of the thermus thermophilus 30s ribosomal subunit2 with a 16s rrna mutation, u20g, bound with streptomycin
44 c1zx4B_
not modelled
5.5
18
PDB header: cell cycleChain: B: PDB Molecule: plasmid partition par b protein;PDBTitle: structure of parb bound to dna
45 d1c0ma2
not modelled
5.4
19
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
46 c3v22U_
not modelled
5.4
47
PDB header: ribosomeChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: crystal structure of rmf bound to the 70s ribosome. this pdb entry2 contains coordinates for the 30s subunit with bound rmf of the 1st3 ribosome in the asu
47 c1n34V_
not modelled
5.4
47
PDB header: ribosomeChain: V: PDB Molecule: 30s ribosomal protein thx;PDBTitle: structure of the thermus thermophilus 30s ribosomal subunit2 in the presence of codon and crystallographically3 disordered near-cognate transfer rna anticodon stem-loop4 mismatched at the first codon position
48 c3ohyU_
not modelled
5.3
47
PDB header: ribosome/antibioticChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: structure of the thermus thermophilus 70s ribosome complexed with2 azithromycin. this file contains the 30s subunit of one 70s ribosome.3 the entire crystal structure contains two 70s ribosomes.
49 c3oi0U_
not modelled
5.3
47
PDB header: ribosome/antibioticChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: structure of the thermus thermophilus 70s ribosome complexed with2 azithromycin. this file contains the 30s subunit of one 70s ribosome.3 the entire crystal structure contains two 70s ribosomes.
50 c2ow8v_
not modelled
5.2
47
PDB header: ribosomeChain: V: PDB Molecule: PDBTitle: crystal structure of a 70s ribosome-trna complex reveals functional2 interactions and rearrangements. this file, 2ow8, contains the 30s3 ribosome subunit, two trna, and mrna molecules. 50s ribosome subunit4 is in the file 1vsa.
51 d1h3za_
not modelled
5.1
10
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: PWWP domain
52 c1i97U_
not modelled
5.1
47
PDB header: ribosomeChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: crystal structure of the 30s ribosomal subunit from thermus2 thermophilus in complex with tetracycline
53 c3otoU_
not modelled
5.0
47
PDB header: ribosomeChain: U: PDB Molecule: 30s ribosomal protein thx;PDBTitle: crystal structure of the 30s ribosomal subunit from a ksga mutant of2 thermus thermophilus (hb8)
54 c4dv5U_
not modelled
5.0
47
PDB header: ribosome/antibioticChain: U: PDB Molecule: ribosomal protein thx;PDBTitle: crystal structure of the thermus thermophilus 30s ribosomal subunit2 with a 16s rrna mutation, a914g, bound with streptomycin