| 1 | c3lu0C_
|
|
 |
100.0 |
58 |
PDB header:transferase
Chain: C: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: molecular model of escherichia coli core rna polymerase
|
|
|
|
| 2 | c6j9eC_
|
|
 |
100.0 |
56 |
PDB header:transcription
Chain: C: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: cryo-em structure of xanthomonos oryzae transcription elongation2 complex with nusa and the bacteriophage protein p7
|
|
|
|
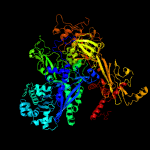
| 3 | c5tw1C_
|
|
 |
100.0 |
93 |
PDB header:transcription activator/transferase/dna
Chain: C: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: crystal structure of a mycobacterium smegmatis transcription2 initiation complex with rbpa
|
|
|
|
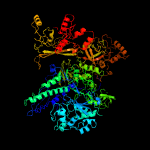
| 4 | d1smyc_
|
|
 |
100.0 |
52 |
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta |
|
|
|
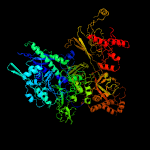
| 5 | d1ynjc1
|
|
 |
100.0 |
51 |
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta |
|
|
|
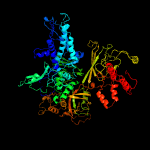
| 6 | c6dcfC_
|
|
 |
100.0 |
95 |
PDB header:transcription/dna/antibiotic
Chain: C: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: crystal structure of a mycobacterium smegmatis transcription2 initiation complex with rifampicin-resistant rna polymerase and bound3 to kanglemycin a
|
|
|
|
| 7 | c4c2mQ_
|
|
 |
100.0 |
24 |
PDB header:transcription
Chain: Q: PDB Molecule:dna-directed rna polymerase i subunit rpa135;
PDBTitle: structure of rna polymerase i at 2.8 a resolution
|
|
|
|
| 8 | c3iydC_
|
|
 |
100.0 |
57 |
PDB header:transcription/dna
Chain: C: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: three-dimensional em structure of an intact activator-dependent2 transcription initiation complex
|
|
|
|
| 9 | c5flmB_
|
|
 |
100.0 |
27 |
PDB header:transcription
Chain: B: PDB Molecule:dna-directed rna polymerase ii subunit rpb2;
PDBTitle: structure of transcribing mammalian rna polymerase ii
|
|
|
|
| 10 | c5fj9B_
|
|
 |
100.0 |
27 |
PDB header:transcription
Chain: B: PDB Molecule:dna-directed rna polymerase iii subunit rpc2;
PDBTitle: cryo-em structure of yeast apo rna polymerase iii at 4.6 a
|
|
|
|
| 11 | c2pmzB_
|
|
 |
100.0 |
31 |
PDB header:translation, transferase
Chain: B: PDB Molecule:dna-directed rna polymerase subunit b;
PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
|
|
|
|
| 12 | c3h0gN_
|
|
 |
100.0 |
27 |
PDB header:transcription
Chain: N: PDB Molecule:dna-directed rna polymerase ii subunit rpb2;
PDBTitle: rna polymerase ii from schizosaccharomyces pombe
|
|
|
|
| 13 | d1twfb_
|
|
 |
100.0 |
27 |
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta |
|
|
|
| 14 | c4qiwB_
|
|
 |
100.0 |
31 |
PDB header:transcription
Chain: B: PDB Molecule:dna-directed rna polymerase;
PDBTitle: crystal structure of euryarchaeal rna polymerase from thermococcus2 kodakarensis
|
|
|
|
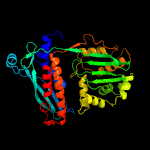
| 15 | c4kbmA_
|
|
 |
100.0 |
99 |
PDB header:transferase/transcription
Chain: A: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: structure of the mtb card/rnap beta subunit b1-b2 domains complex
|
|
|
|
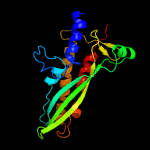
| 16 | c3mlqD_
|
|
 |
100.0 |
55 |
PDB header:transferase/transcription
Chain: D: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: crystal structure of the thermus thermophilus transcription-repair2 coupling factor rna polymerase interacting domain with the thermus3 aquaticus rna polymerase beta1 domain
|
|
|
|
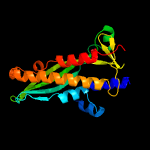
| 17 | c3mlqB_
|
|
 |
100.0 |
44 |
PDB header:transferase/transcription
Chain: B: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: crystal structure of the thermus thermophilus transcription-repair2 coupling factor rna polymerase interacting domain with the thermus3 aquaticus rna polymerase beta1 domain
|
|
|
|
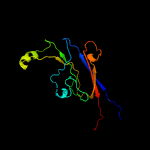
| 18 | c2ly7A_
|
|
 |
100.0 |
74 |
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: b-flap domain of rna polymerase (b. subtilis)
|
|
|
|
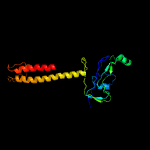
| 19 | c3tbiB_
|
|
 |
100.0 |
44 |
PDB header:transcription
Chain: B: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: crystal structure of t4 gp33 bound to e. coli rnap beta-flap domain
|
|
|
|
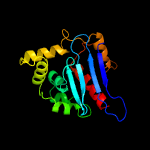
| 20 | c3ltiA_
|
|
 |
100.0 |
50 |
PDB header:transferase
Chain: A: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: crystal structure of the escherichia coli rna polymerase beta subunit2 beta2-betai4 domains
|
|
|
|
| 21 | c3qqcA_ |
|
not modelled |
99.4 |
22 |
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase subunit b, dna-directed rna
PDBTitle: crystal structure of archaeal spt4/5 bound to the rnap clamp domain
|
|
|
| 22 | c5j1lA_ |
|
not modelled |
93.5 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:toxr-activated gene (tage);
PDBTitle: crystal structure of csd1-csd2 dimer i
|
|
|
| 23 | c2lmcB_ |
|
not modelled |
92.8 |
27 |
PDB header:transcription
Chain: B: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: structure of t7 transcription factor gp2-e. coli rnap jaw domain2 complex
|
|
|
| 24 | c2gu1A_ |
|
not modelled |
89.9 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:zinc peptidase;
PDBTitle: crystal structure of a zinc containing peptidase from2 vibrio cholerae
|
|
|
| 25 | c2hsiB_ |
|
not modelled |
89.3 |
26 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative peptidase m23;
PDBTitle: crystal structure of putative peptidase m23 from2 pseudomonas aeruginosa, new york structural genomics3 consortium
|
|
|
| 26 | c3sluB_ |
|
not modelled |
84.0 |
27 |
PDB header:hydrolase
Chain: B: PDB Molecule:m23 peptidase domain protein;
PDBTitle: crystal structure of nmb0315
|
|
|
| 27 | c5j1mD_ |
|
not modelled |
80.6 |
23 |
PDB header:hydrolase
Chain: D: PDB Molecule:toxr-activated gene (tage);
PDBTitle: crystal structure of csd1-csd2 dimer ii
|
|
|
| 28 | d1qwya_ |
|
not modelled |
80.4 |
31 |
Fold:Barrel-sandwich hybrid
Superfamily:Duplicated hybrid motif
Family:Peptidoglycan hydrolase LytM |
|
|
| 29 | c4bh5B_ |
|
not modelled |
79.7 |
33 |
PDB header:cell cycle
Chain: B: PDB Molecule:murein hydrolase activator envc;
PDBTitle: lytm domain of envc, an activator of cell wall amidases in2 escherichia coli
|
|
|
| 30 | c4rnzA_ |
|
not modelled |
74.6 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:conserved hypothetical secreted protein;
PDBTitle: structure of helicobacter pylori csd3 from the hexagonal crystal
|
|
|
| 31 | c5gt1A_ |
|
not modelled |
73.9 |
24 |
PDB header:choline-binding protein
Chain: A: PDB Molecule:choline binding protein a;
PDBTitle: crystal structure of cbpa from l. salivarius ren
|
|
|
| 32 | c2aujD_ |
|
not modelled |
73.4 |
25 |
PDB header:transferase
Chain: D: PDB Molecule:dna-directed rna polymerase beta' chain;
PDBTitle: structure of thermus aquaticus rna polymerase beta'-subunit2 insert
|
|
|
| 33 | c4lxcA_ |
|
not modelled |
73.1 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:lysostaphin;
PDBTitle: the antimicrobial peptidase lysostaphin from staphylococcus simulans
|
|
|
| 34 | c4qpbB_ |
|
not modelled |
71.5 |
32 |
PDB header:hydrolase
Chain: B: PDB Molecule:lysostaphin;
PDBTitle: catalytic domain of the antimicrobial peptidase lysostaphin from2 staphylococcus simulans crystallized in the absence of phosphate
|
|
|
| 35 | c3tufB_ |
|
not modelled |
71.3 |
30 |
PDB header:signaling protein
Chain: B: PDB Molecule:stage ii sporulation protein q;
PDBTitle: structure of the spoiiq-spoiiiah pore forming complex.
|
|
|
| 36 | c5kvpA_ |
|
not modelled |
68.4 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:zoocin a endopeptidase;
PDBTitle: solution structure of the catalytic domain of zoocin a
|
|
|
| 37 | c2b44A_ |
|
not modelled |
64.3 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycyl-glycine endopeptidase lytm;
PDBTitle: truncated s. aureus lytm, p 32 2 1 crystal form
|
|
|
| 38 | c3uz0D_ |
|
not modelled |
61.5 |
29 |
PDB header:transport protein
Chain: D: PDB Molecule:stage ii sporulation protein q;
PDBTitle: crystal structure of spoiiiah and spoiiq complex
|
|
|
| 39 | c2aukA_ |
|
not modelled |
56.6 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:dna-directed rna polymerase beta' chain;
PDBTitle: structure of e. coli rna polymerase beta' g/g' insert
|
|
|
| 40 | d1r46a1 |
|
not modelled |
54.4 |
24 |
Fold:Glycosyl hydrolase domain
Superfamily:Glycosyl hydrolase domain
Family:alpha-Amylases, C-terminal beta-sheet domain |
|
|
| 41 | c6ekaB_ |
|
not modelled |
47.1 |
86 |
PDB header:protein fibril
Chain: B: PDB Molecule:podospora anserina s mat+ genomic dna chromosome 3,
PDBTitle: solid-state mas nmr structure of the hellf prion amyloid fibrils
|
|
|
| 42 | c5kqbA_ |
|
not modelled |
47.0 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidase m23;
PDBTitle: identification and structural characterization of lytu
|
|
|
| 43 | c5b0hB_ |
|
not modelled |
44.6 |
11 |
PDB header:metal binding protein
Chain: B: PDB Molecule:leukocyte cell-derived chemotaxin-2;
PDBTitle: crystal structure of human leukocyte cell-derived chemotaxin 2
|
|
|
| 44 | c3nyyA_ |
|
not modelled |
44.6 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative glycyl-glycine endopeptidase lytm;
PDBTitle: crystal structure of a putative glycyl-glycine endopeptidase lytm2 (rumgna_02482) from ruminococcus gnavus atcc 29149 at 1.60 a3 resolution
|
|
|
| 45 | c4yk3B_ |
|
not modelled |
42.1 |
30 |
PDB header:protein binding
Chain: B: PDB Molecule:bepe protein;
PDBTitle: crystal structure of the bid domain of bepe from bartonella henselae
|
|
|
| 46 | c4yk2B_ |
|
not modelled |
42.1 |
20 |
PDB header:protein binding
Chain: B: PDB Molecule:bartonella effector protein (bep) substrate of virb t4ss;
PDBTitle: crystal structure of the bid domain of bep9 from bartonella2 clarridgeiae
|
|
|
| 47 | d2gpra_ |
|
not modelled |
42.1 |
16 |
Fold:Barrel-sandwich hybrid
Superfamily:Duplicated hybrid motif
Family:Glucose permease-like |
|
|
| 48 | d1glaf_ |
|
not modelled |
40.3 |
20 |
Fold:Barrel-sandwich hybrid
Superfamily:Duplicated hybrid motif
Family:Glucose permease-like |
|
|
| 49 | d2f3ga_ |
|
not modelled |
36.5 |
20 |
Fold:Barrel-sandwich hybrid
Superfamily:Duplicated hybrid motif
Family:Glucose permease-like |
|
|
| 50 | d1gpra_ |
|
not modelled |
35.9 |
28 |
Fold:Barrel-sandwich hybrid
Superfamily:Duplicated hybrid motif
Family:Glucose permease-like |
|
|
| 51 | c2ivfC_ |
|
not modelled |
35.0 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:ethylbenzene dehydrogenase gamma-subunit;
PDBTitle: ethylbenzene dehydrogenase from aromatoleum aromaticum
|
|
|
| 52 | d1uc8a1 |
|
not modelled |
32.3 |
18 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:Lysine biosynthesis enzyme LysX, N-terminal domain |
|
|
| 53 | c2gtiA_ |
|
not modelled |
32.2 |
16 |
PDB header:viral protein
Chain: A: PDB Molecule:replicase polyprotein 1ab;
PDBTitle: mutation of mhv coronavirus non-structural protein nsp15 (f307l)
|
|
|
| 54 | c4yk1A_ |
|
not modelled |
29.7 |
10 |
PDB header:protein binding
Chain: A: PDB Molecule:bartonella effector protein (bep) substrate of virb t4ss;
PDBTitle: crystal structure of the bid domain of bep6 from bartonella rochalimae
|
|
|
| 55 | c1wcoN_ |
|
not modelled |
29.1 |
86 |
PDB header:peptide/antibiotic
Chain: N: PDB Molecule:nisin z;
PDBTitle: the solution structure of the nisin-lipid ii complex
|
|
|
| 56 | d1y4oa1 |
|
not modelled |
28.8 |
53 |
Fold:Profilin-like
Superfamily:Roadblock/LC7 domain
Family:Roadblock/LC7 domain |
|
|
| 57 | c6h5nA_ |
|
not modelled |
27.8 |
27 |
PDB header:cell invasion
Chain: A: PDB Molecule:gametocyte surface protein p45/48;
PDBTitle: plasmodium falciparum pfs48/45 c-terminal domain bound to monoclonal2 antibody 85rf45.1
|
|
|
| 58 | d1n26a2 |
|
not modelled |
26.6 |
14 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Fibronectin type III
Family:Fibronectin type III |
|
|
| 59 | c4p6vA_ |
|
not modelled |
25.8 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:na(+)-translocating nadh-quinone reductase subunit a;
PDBTitle: crystal structure of the na+-translocating nadh: ubiquinone2 oxidoreductase from vibrio cholerae
|
|
|
| 60 | d1gm5a4 |
|
not modelled |
25.6 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Tandem AAA-ATPase domain |
|
|
| 61 | c2db3D_ |
|
not modelled |
24.8 |
33 |
PDB header:hydrolase/rna
Chain: D: PDB Molecule:atp-dependent rna helicase vasa;
PDBTitle: structural basis for rna unwinding by the dead-box protein2 drosophila vasa
|
|
|
| 62 | c4iqzD_ |
|
not modelled |
24.2 |
16 |
PDB header:unknown function
Chain: D: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: the crystal structure of a large insert in rna polymerase (rpoc)2 subunit from e. coli
|
|
|
| 63 | d1ni7a_ |
|
not modelled |
23.8 |
18 |
Fold:SufE/NifU
Superfamily:SufE/NifU
Family:SufE-like |
|
|
| 64 | c4nhoA_ |
|
not modelled |
22.5 |
38 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable atp-dependent rna helicase ddx23;
PDBTitle: structure of the spliceosomal dead-box protein prp28
|
|
|
| 65 | d1lk5a2 |
|
not modelled |
22.4 |
17 |
Fold:Ferredoxin-like
Superfamily:D-ribose-5-phosphate isomerase (RpiA), lid domain
Family:D-ribose-5-phosphate isomerase (RpiA), lid domain |
|
|
| 66 | d2eyqa5 |
|
not modelled |
22.3 |
16 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Tandem AAA-ATPase domain |
|
|
| 67 | d1mzga_ |
|
not modelled |
21.8 |
14 |
Fold:SufE/NifU
Superfamily:SufE/NifU
Family:SufE-like |
|
|
| 68 | c4meiA_ |
|
not modelled |
21.2 |
16 |
PDB header:protein transport
Chain: A: PDB Molecule:virb8 protein;
PDBTitle: crystal structure of a virb8 type iv secretion system machinery2 soluble domain from bartonella tribocorum
|
|
|
| 69 | c5m88A_ |
|
not modelled |
21.0 |
23 |
PDB header:splicing
Chain: A: PDB Molecule:core;
PDBTitle: spliceosome component
|
|
|
| 70 | c5c22A_ |
|
not modelled |
21.0 |
53 |
PDB header:protein transport
Chain: A: PDB Molecule:chromosomal hemolysin d;
PDBTitle: crystal structure of zn-bound hlyd from e. coli
|
|
|
| 71 | c4bmjC_ |
|
not modelled |
20.4 |
50 |
PDB header:apoptosis
Chain: C: PDB Molecule:tax1-binding protein 1;
PDBTitle: structure of the ubz1and2 tandem of the ubiquitin-binding adaptor2 protein tax1bp1
|
|
|
| 72 | c4l0zA_ |
|
not modelled |
20.4 |
21 |
PDB header:transcription/dna
Chain: A: PDB Molecule:runt-related transcription factor 1;
PDBTitle: crystal structure of runx1 and ets1 bound to tcr alpha promoter2 (crystal form 2)
|
|
|
| 73 | c1rb8J_ |
|
not modelled |
20.2 |
55 |
PDB header:virus/dna
Chain: J: PDB Molecule:small core protein;
PDBTitle: the phix174 dna binding protein j in two different capsid2 environments.
|
|
|
| 74 | c3mjhD_ |
|
not modelled |
19.9 |
50 |
PDB header:protein transport
Chain: D: PDB Molecule:early endosome antigen 1;
PDBTitle: crystal structure of human rab5a in complex with the c2h2 zinc finger2 of eea1
|
|
|
| 75 | c3bxzA_ |
|
not modelled |
19.7 |
39 |
PDB header:transport protein
Chain: A: PDB Molecule:preprotein translocase subunit seca;
PDBTitle: crystal structure of the isolated dead motor domains from escherichia2 coli seca
|
|
|
| 76 | c4upcA_ |
|
not modelled |
19.7 |
20 |
PDB header:protein binding
Chain: A: PDB Molecule:nicastrin;
PDBTitle: structure of a extracellular domain
|
|
|
| 77 | c5xyiT_ |
|
not modelled |
19.5 |
20 |
PDB header:ribosome
Chain: T: PDB Molecule:ribosomal protein s19e, putative;
PDBTitle: small subunit of trichomonas vaginalis ribosome
|
|
|
| 78 | c2vb0A_ |
|
not modelled |
19.2 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:polyprotein 3bcd;
PDBTitle: crystal structure of coxsackievirus b3 proteinase 3c
|
|
|
| 79 | c2nu9E_ |
|
not modelled |
18.3 |
21 |
PDB header:ligase
Chain: E: PDB Molecule:succinyl-coa synthetase beta chain;
PDBTitle: c123at mutant of e. coli succinyl-coa synthetase2 orthorhombic crystal form
|
|
|
| 80 | d1cf2o2 |
|
not modelled |
18.3 |
29 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
|
|
| 81 | c5xxuI_ |
|
not modelled |
17.3 |
35 |
PDB header:ribosome
Chain: I: PDB Molecule:ribosomal protein es8;
PDBTitle: small subunit of toxoplasma gondii ribosome
|
|
|
| 82 | c5gmkq_ |
|
not modelled |
17.2 |
24 |
PDB header:rna binding protein/rna
Chain: Q: PDB Molecule:pre-mrna-splicing factor slt11;
PDBTitle: cryo-em structure of the catalytic step i spliceosome (c complex) at2 3.4 angstrom resolution
|
|
|
| 83 | c3it5B_ |
|
not modelled |
17.0 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:protease lasa;
PDBTitle: crystal structure of the lasa virulence factor from pseudomonas2 aeruginosa
|
|
|
| 84 | c3cwiA_ |
|
not modelled |
16.8 |
18 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:thiamine-biosynthesis protein this;
PDBTitle: crystal structure of thiamine biosynthesis protein (this)2 from geobacter metallireducens. northeast structural3 genomics consortium target gmr137
|
|
|
| 85 | d1tf5a4 |
|
not modelled |
15.8 |
39 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Tandem AAA-ATPase domain |
|
|
| 86 | d1d02a_ |
|
not modelled |
15.5 |
17 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:Restriction endonuclease MunI |
|
|
| 87 | d1px5a1 |
|
not modelled |
15.3 |
28 |
Fold:PAP/OAS1 substrate-binding domain
Superfamily:PAP/OAS1 substrate-binding domain
Family:2'-5'-oligoadenylate synthetase 1, OAS1, second domain |
|
|
| 88 | c6em5b_ |
|
not modelled |
15.3 |
25 |
PDB header:ribosome
Chain: B: PDB Molecule:60s ribosomal protein l3;
PDBTitle: state d architectural model (nsa1-tap flag-ytm1) - visualizing the2 assembly pathway of nucleolar pre-60s ribosomes
|
|
|
| 89 | c6g2jY_ |
|
not modelled |
15.2 |
38 |
PDB header:oxidoreductase
Chain: Y: PDB Molecule:mcg5603;
PDBTitle: mouse mitochondrial complex i in the active state
|
|
|
| 90 | c4nhfF_ |
|
not modelled |
15.2 |
17 |
PDB header:protein transport
Chain: F: PDB Molecule:trwg protein;
PDBTitle: crystal structure of the soluble domain of trwg type iv secretion2 machinery from bartonella grahamii
|
|
|
| 91 | c3izbX_ |
|
not modelled |
15.2 |
42 |
PDB header:ribosome
Chain: X: PDB Molecule:40s ribosomal protein rps27 (s27e);
PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
|
|
| 92 | c3j20W_ |
|
not modelled |
15.1 |
33 |
PDB header:ribosome
Chain: W: PDB Molecule:30s ribosomal protein s27e;
PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (30s3 ribosomal subunit)
|
|
|
| 93 | c1gjjA_ |
|
not modelled |
15.0 |
35 |
PDB header:membrane protein
Chain: A: PDB Molecule:lap2;
PDBTitle: n-terminal constant region of the nuclear envelope protein2 lap2
|
|
|
| 94 | c2qhoB_ |
|
not modelled |
14.9 |
52 |
PDB header:protein binding/ligase
Chain: B: PDB Molecule:e3 ubiquitin-protein ligase edd1;
PDBTitle: crystal structure of the uba domain from edd ubiquitin2 ligase in complex with ubiquitin
|
|
|
| 95 | c1m1jA_ |
|
not modelled |
14.9 |
50 |
PDB header:blood clotting
Chain: A: PDB Molecule:fibrinogen alpha subunit;
PDBTitle: crystal structure of native chicken fibrinogen with two different2 bound ligands
|
|
|
| 96 | c2f1mA_ |
|
not modelled |
14.8 |
20 |
PDB header:transport protein
Chain: A: PDB Molecule:acriflavine resistance protein a;
PDBTitle: conformational flexibility in the multidrug efflux system protein acra
|
|
|
| 97 | c3wfoB_ |
|
not modelled |
14.8 |
34 |
PDB header:transferase
Chain: B: PDB Molecule:poly a polymerase;
PDBTitle: trna processing enzyme (apo form 1)
|
|
|
| 98 | c1bplA_ |
|
not modelled |
14.6 |
26 |
PDB header:glycosyltransferase
Chain: A: PDB Molecule:alpha-1,4-glucan-4-glucanohydrolase;
PDBTitle: glycosyltransferase
|
|
|
| 99 | c2bhmE_ |
|
not modelled |
14.6 |
8 |
PDB header:bacterial protein
Chain: E: PDB Molecule:type iv secretion system protein virb8;
PDBTitle: crystal structure of virb8 from brucella suis
|
|
|