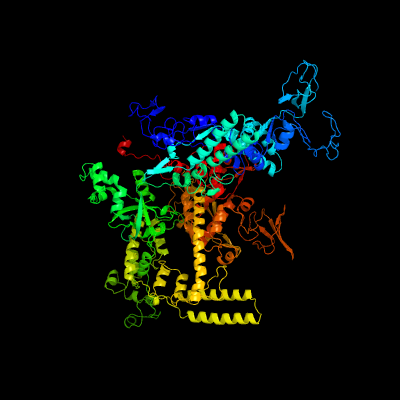
| 1 | c4g7oN_
|
|
 |
100.0 |
49 |
PDB header:transcription, transferase/dna
Chain: N: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: crystal structure of thermus thermophilus transcription initiation2 complex containing 2 nt of rna
|
|
|
|
| 2 | d1smyd_
|
|
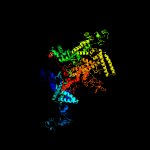 |
100.0 |
48 |
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta-prime |
|
|
|
| 3 | c5tw1D_
|
|
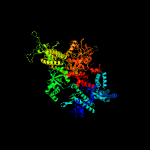 |
100.0 |
87 |
PDB header:transcription activator/transferase/dna
Chain: D: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: crystal structure of a mycobacterium smegmatis transcription2 initiation complex with rbpa
|
|
|
|
| 4 | c2o5jN_
|
|
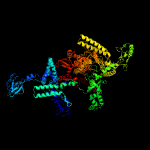 |
100.0 |
49 |
PDB header:transferase/dna-rna hybrid
Chain: N: PDB Molecule:dna-directed rna polymerase beta' chain;
PDBTitle: crystal structure of the t. thermophilus rnap polymerase elongation2 complex with the ntp substrate analog
|
|
|
|
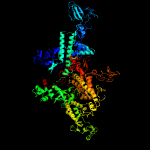
| 5 | c5x22D_
|
|
 |
100.0 |
52 |
PDB header:transferase/dna
Chain: D: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: crystal structure of thermus thermophilus transcription initiation2 complex with gpa and cmpcpp
|
|
|
|
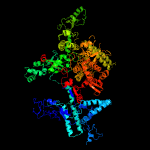
| 6 | c4wqsD_
|
|
 |
100.0 |
53 |
PDB header:transferase/dna/rna
Chain: D: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: thermus thermophilus rna polymerase backtracked complex
|
|
|
|
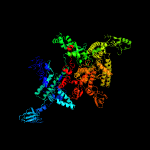
| 7 | c2o5iD_
|
|
 |
100.0 |
48 |
PDB header:transferase/dna-rna hybrid
Chain: D: PDB Molecule:dna-directed rna polymerase beta' chain;
PDBTitle: crystal structure of the t. thermophilus rna polymerase elongation2 complex
|
|
|
|
| 8 | c5xj0D_
|
|
 |
100.0 |
49 |
PDB header:transferase/transcription
Chain: D: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: t. thermophilus rna polymerase holoenzyme bound with gp39 and gp76
|
|
|
|
| 9 | c6cceD_
|
|
 |
100.0 |
85 |
PDB header:transcription/dna/antibiotic
Chain: D: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: crystal structure of a mycobacterium smegmatis rna polymerase2 transcription initiation complex with inhibitor kanglemycin a
|
|
|
|
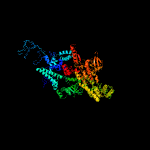
| 10 | c3aoiN_
|
|
 |
100.0 |
47 |
PDB header:transcription, transferase/dna/rna
Chain: N: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: rna polymerase-gfh1 complex (crystal type 2)
|
|
|
|
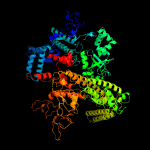
| 11 | c4xsxJ_
|
|
 |
100.0 |
49 |
PDB header:transcription/antibiotic
Chain: J: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: crystal structure of cbr 703 bound to escherichia coli rna polymerase2 holoenzyme
|
|
|
|
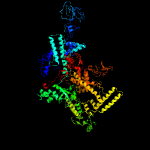
| 12 | c3aoiI_
|
|
 |
100.0 |
47 |
PDB header:transcription, transferase/dna/rna
Chain: I: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: rna polymerase-gfh1 complex (crystal type 2)
|
|
|
|
| 13 | c1l9uD_
|
|
 |
100.0 |
49 |
PDB header:transcription
Chain: D: PDB Molecule:rna polymerase, beta-prime subunit;
PDBTitle: thermus aquaticus rna polymerase holoenzyme at 4 a2 resolution
|
|
|
|
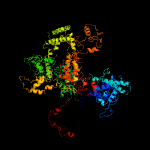
| 14 | c3h0gA_
|
|
 |
100.0 |
23 |
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase ii subunit rpb1;
PDBTitle: rna polymerase ii from schizosaccharomyces pombe
|
|
|
|
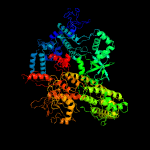
| 15 | c6j9eD_
|
|
 |
100.0 |
52 |
PDB header:transcription
Chain: D: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: cryo-em structure of xanthomonos oryzae transcription elongation2 complex with nusa and the bacteriophage protein p7
|
|
|
|
| 16 | c4igcD_
|
|
 |
100.0 |
51 |
PDB header:transcription, transferase
Chain: D: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: x-ray crystal structure of escherichia coli sigma70 holoenzyme
|
|
|
|
| 17 | c3iydD_
|
|
 |
100.0 |
48 |
PDB header:transcription/dna
Chain: D: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: three-dimensional em structure of an intact activator-dependent2 transcription initiation complex
|
|
|
|
| 18 | c1i6vD_
|
|
 |
100.0 |
49 |
PDB header:transcription
Chain: D: PDB Molecule:dna-directed rna polymerase;
PDBTitle: thermus aquaticus core rna polymerase-rifampicin complex
|
|
|
|
| 19 | d1i6vd_
|
|
 |
100.0 |
49 |
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta-prime |
|
|
|
| 20 | c6gmlA_
|
|
 |
100.0 |
25 |
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase subunit;
PDBTitle: structure of paused transcription complex pol ii-dsif-nelf
|
|
|
|
| 21 | c6dcfD_ |
|
not modelled |
100.0 |
86 |
PDB header:transcription/dna/antibiotic
Chain: D: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: crystal structure of a mycobacterium smegmatis transcription2 initiation complex with rifampicin-resistant rna polymerase and bound3 to kanglemycin a
|
|
|
| 22 | d1twfa_ |
|
not modelled |
100.0 |
25 |
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta-prime |
|
|
| 23 | c4llgJ_ |
|
not modelled |
100.0 |
52 |
PDB header:transferase
Chain: J: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: crystal structure analysis of the e.coli holoenzyme/gp2 complex
|
|
|
| 24 | c5flmA_ |
|
not modelled |
100.0 |
26 |
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase;
PDBTitle: structure of transcribing mammalian rna polymerase ii
|
|
|
| 25 | c5tbzD_ |
|
not modelled |
100.0 |
52 |
PDB header:transcription/rna
Chain: D: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: e. coli rna polymerase complexed with nusg
|
|
|
| 26 | c4c3iA_ |
|
not modelled |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:dna-directed rna polymerase i subunit rpa190;
PDBTitle: structure of 14-subunit rna polymerase i at 3.0 a resolution, crystal2 form c2-100
|
|
|
| 27 | c5m3fA_ |
|
not modelled |
100.0 |
21 |
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase i subunit rpa190;
PDBTitle: yeast rna polymerase i elongation complex at 3.8a
|
|
|
| 28 | c1i6hA_ |
|
not modelled |
100.0 |
25 |
PDB header:transcription/dna-rna hybrid
Chain: A: PDB Molecule:dna-directed rna polymerase ii largest subunit;
PDBTitle: rna polymerase ii elongation complex
|
|
|
| 29 | c5fjaA_ |
|
not modelled |
100.0 |
22 |
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase iii subunit rpc1;
PDBTitle: cryo-em structure of yeast rna polymerase iii at 4.7 a
|
|
|
| 30 | d1ynjd1 |
|
not modelled |
100.0 |
48 |
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta-prime |
|
|
| 31 | c1twaA_ |
|
not modelled |
100.0 |
24 |
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase ii largest subunit;
PDBTitle: rna polymerase ii complexed with atp
|
|
|
| 32 | c5lmxA_ |
|
not modelled |
100.0 |
20 |
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase i subunit rpa190;
PDBTitle: monomeric rna polymerase i at 4.9 a resolution
|
|
|
| 33 | c4b1pW_ |
|
not modelled |
100.0 |
27 |
PDB header:transferase/dna
Chain: W: PDB Molecule:dna-directed rna polymerase;
PDBTitle: archaeal rnap-dna binary complex at 4.32ang
|
|
|
| 34 | c4qiwA_ |
|
not modelled |
100.0 |
26 |
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase;
PDBTitle: crystal structure of euryarchaeal rna polymerase from thermococcus2 kodakarensis
|
|
|
| 35 | c2pmzQ_ |
|
not modelled |
100.0 |
31 |
PDB header:translation, transferase
Chain: Q: PDB Molecule:dna-directed rna polymerase subunit a;
PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
|
|
|
| 36 | c6drdA_ |
|
not modelled |
100.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:dna-directed rna polymerase ii subunit rpb1;
PDBTitle: rna pol ii(g)
|
|
|
| 37 | c3qqcA_ |
|
not modelled |
100.0 |
28 |
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase subunit b, dna-directed rna
PDBTitle: crystal structure of archaeal spt4/5 bound to the rnap clamp domain
|
|
|
| 38 | c4qiwC_ |
|
not modelled |
100.0 |
24 |
PDB header:transcription
Chain: C: PDB Molecule:dna-directed rna polymerase subunit a'';
PDBTitle: crystal structure of euryarchaeal rna polymerase from thermococcus2 kodakarensis
|
|
|
| 39 | c2wb1Y_ |
|
not modelled |
100.0 |
19 |
PDB header:transcription
Chain: Y: PDB Molecule:dna-directed rna polymerase rpo1c subunit;
PDBTitle: the complete structure of the archaeal 13-subunit dna-2 directed rna polymerase
|
|
|
| 40 | c2pmzG_ |
|
not modelled |
100.0 |
24 |
PDB header:translation, transferase
Chain: G: PDB Molecule:dna-directed rna polymerase subunit a";
PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
|
|
|
| 41 | c2aujD_ |
|
not modelled |
96.6 |
14 |
PDB header:transferase
Chain: D: PDB Molecule:dna-directed rna polymerase beta' chain;
PDBTitle: structure of thermus aquaticus rna polymerase beta'-subunit2 insert
|
|
|
| 42 | c3e7hA_ |
|
not modelled |
96.4 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: the crystal structure of the beta subunit of the dna-directed rna2 polymerase from vibrio cholerae o1 biovar eltor
|
|
|
| 43 | c2lmcB_ |
|
not modelled |
95.8 |
16 |
PDB header:transcription
Chain: B: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: structure of t7 transcription factor gp2-e. coli rnap jaw domain2 complex
|
|
|
| 44 | c2aukA_ |
|
not modelled |
93.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:dna-directed rna polymerase beta' chain;
PDBTitle: structure of e. coli rna polymerase beta' g/g' insert
|
|
|
| 45 | c5fswC_ |
|
not modelled |
89.0 |
29 |
PDB header:transferase
Chain: C: PDB Molecule:rna dependent rna polymerase qde-1;
PDBTitle: rna dependent rna polymerase qde-1 from thielavia terrestris
|
|
|
| 46 | c2j7nA_ |
|
not modelled |
88.7 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:rna-dependent rna polymerase;
PDBTitle: structure of the rnai polymerase from neurospora crassa
|
|
|
| 47 | c3mkrB_ |
|
not modelled |
80.1 |
29 |
PDB header:transport protein
Chain: B: PDB Molecule:coatomer subunit alpha;
PDBTitle: crystal structure of yeast alpha/epsilon-cop subcomplex of the copi2 vesicular coat
|
|
|
| 48 | c3mv2A_ |
|
not modelled |
74.3 |
15 |
PDB header:protein transport
Chain: A: PDB Molecule:coatomer subunit alpha;
PDBTitle: crystal structure of a-cop in complex with e-cop
|
|
|
| 49 | d1odha_ |
|
not modelled |
68.4 |
29 |
Fold:GCM domain
Superfamily:GCM domain
Family:GCM domain |
|
|
| 50 | c4iqzD_ |
|
not modelled |
63.1 |
21 |
PDB header:unknown function
Chain: D: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: the crystal structure of a large insert in rna polymerase (rpoc)2 subunit from e. coli
|
|
|
| 51 | c4zlhB_ |
|
not modelled |
54.9 |
16 |
PDB header:metal binding protein
Chain: B: PDB Molecule:lipopolysaccharide assembly protein b;
PDBTitle: structure of the lapb cytoplasmic domain at 2 angstroms
|
|
|
| 52 | c2lcqA_ |
|
not modelled |
50.6 |
30 |
PDB header:metal binding protein
Chain: A: PDB Molecule:putative toxin vapc6;
PDBTitle: solution structure of the endonuclease nob1 from p.horikoshii
|
|
|
| 53 | c3hcjB_ |
|
not modelled |
48.4 |
42 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:peptide methionine sulfoxide reductase;
PDBTitle: structure of msrb from xanthomonas campestris (oxidized2 form)
|
|
|
| 54 | d1xm0a1 |
|
not modelled |
47.7 |
17 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:SelR domain |
|
|
| 55 | c3ky9B_ |
|
not modelled |
45.6 |
25 |
PDB header:apoptosis
Chain: B: PDB Molecule:proto-oncogene vav;
PDBTitle: autoinhibited vav1
|
|
|
| 56 | c2l1uA_ |
|
not modelled |
44.7 |
42 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methionine-r-sulfoxide reductase b2, mitochondrial;
PDBTitle: structure-functional analysis of mammalian msrb2 protein
|
|
|
| 57 | c3cezA_ |
|
not modelled |
44.4 |
42 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methionine-r-sulfoxide reductase;
PDBTitle: crystal structure of methionine-r-sulfoxide reductase from2 burkholderia pseudomallei
|
|
|
| 58 | c2k8dA_ |
|
not modelled |
43.8 |
42 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:peptide methionine sulfoxide reductase msrb;
PDBTitle: solution structure of a zinc-binding methionine sulfoxide reductase
|
|
|
| 59 | c6g5iy_ |
|
not modelled |
43.1 |
29 |
PDB header:ribosome
Chain: Y: PDB Molecule:40s ribosomal protein s24;
PDBTitle: cryo-em structure of a late human pre-40s ribosomal subunit - state r
|
|
|
| 60 | c2hr5B_ |
|
not modelled |
42.4 |
27 |
PDB header:metal binding protein
Chain: B: PDB Molecule:rubrerythrin;
PDBTitle: pf1283- rubrerythrin from pyrococcus furiosus iron bound form
|
|
|
| 61 | c3e0mB_ |
|
not modelled |
42.1 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:peptide methionine sulfoxide reductase msra/msrb
PDBTitle: crystal structure of fusion protein of msra and msrb
|
|
|
| 62 | d1l1da_ |
|
not modelled |
41.9 |
17 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:SelR domain |
|
|
| 63 | c5a1vK_ |
|
not modelled |
41.6 |
26 |
PDB header:transport protein
Chain: K: PDB Molecule:coatomer subunit alpha;
PDBTitle: the structure of the copi coat linkage i
|
|
|
| 64 | c2gj2A_ |
|
not modelled |
39.3 |
29 |
PDB header:metal binding protein
Chain: A: PDB Molecule:wsv230;
PDBTitle: crystal structure of vp9 from white spot syndrome virus
|
|
|
| 65 | c5fa9B_ |
|
not modelled |
39.2 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:peptide methionine sulfoxide reductase msra;
PDBTitle: bifunctional methionine sulfoxide reductase ab (msrab) from treponema2 denticola
|
|
|
| 66 | c1yuzB_ |
|
not modelled |
38.2 |
30 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nigerythrin;
PDBTitle: partially reduced state of nigerythrin
|
|
|
| 67 | c3t6pA_ |
|
not modelled |
36.9 |
25 |
PDB header:apoptosis
Chain: A: PDB Molecule:baculoviral iap repeat-containing protein 2;
PDBTitle: iap antagonist-induced conformational change in ciap1 promotes e32 ligase activation via dimerization
|
|
|
| 68 | c3uc9A_ |
|
not modelled |
35.3 |
38 |
PDB header:recombination
Chain: A: PDB Molecule:increased recombination centers protein 6;
PDBTitle: crystal structure of yeast irc6p - a novel type of conserved clathrin2 accessory protein
|
|
|
| 69 | c2vrwB_ |
|
not modelled |
34.8 |
24 |
PDB header:signaling protein
Chain: B: PDB Molecule:proto-oncogene vav;
PDBTitle: critical structural role for the ph and c1 domains of the2 vav1 exchange factor
|
|
|
| 70 | c2bx9J_ |
|
not modelled |
34.6 |
46 |
PDB header:transcription regulation
Chain: J: PDB Molecule:tryptophan rna-binding attenuator protein-inhibitory
PDBTitle: crystal structure of b.subtilis anti-trap protein, an2 antagonist of trap-rna interactions
|
|
|
| 71 | c5zb8B_ |
|
not modelled |
34.5 |
25 |
PDB header:dna binding protein
Chain: B: PDB Molecule:pfuendoq;
PDBTitle: crystal structure of the novel lesion-specific endonuclease pfuendoq2 from pyrococcus furiosus
|
|
|
| 72 | d2gmga1 |
|
not modelled |
33.4 |
25 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:PF0610-like |
|
|
| 73 | c2kaoA_ |
|
not modelled |
33.1 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methionine-r-sulfoxide reductase b1;
PDBTitle: structure of reduced mouse methionine sulfoxide reductase b12 (sec95cys mutant)
|
|
|
| 74 | d1x64a2 |
|
not modelled |
32.7 |
20 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
|
|
| 75 | d2eppa1 |
|
not modelled |
31.0 |
36 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
|
|
| 76 | d2g9ha2 |
|
not modelled |
30.2 |
71 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
|
|
| 77 | c4u3eA_ |
|
not modelled |
30.1 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribonucleoside triphosphate reductase;
PDBTitle: anaerobic ribonucleotide reductase
|
|
|
| 78 | c4rasC_ |
|
not modelled |
29.9 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:oxidoreductase, nad-binding/iron-sulfur cluster-binding
PDBTitle: reductive dehalogenase structure suggests a mechanism for b12-2 dependent dehalogenation
|
|
|
| 79 | c5ey2A_ |
|
not modelled |
29.5 |
21 |
PDB header:transcription
Chain: A: PDB Molecule:gtp-sensing transcriptional pleiotropic repressor cody;
PDBTitle: crystal structure of cody from bacillus cereus
|
|
|
| 80 | c2eppA_ |
|
not modelled |
29.3 |
36 |
PDB header:transcription
Chain: A: PDB Molecule:poz-, at hook-, and zinc finger-containing
PDBTitle: solution structure of the first c2h2 type zinc finger2 domain of zinc finger protein 278
|
|
|
| 81 | d1s9va2 |
|
not modelled |
29.1 |
57 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
|
|
| 82 | c1q68B_ |
|
not modelled |
28.8 |
30 |
PDB header:membrane protein/transferase
Chain: B: PDB Molecule:proto-oncogene tyrosine-protein kinase lck;
PDBTitle: solution structure of t-cell surface glycoprotein cd4 and2 proto-oncogene tyrosine-protein kinase lck fragments
|
|
|
| 83 | c1v9pB_ |
|
not modelled |
28.1 |
16 |
PDB header:ligase
Chain: B: PDB Molecule:dna ligase;
PDBTitle: crystal structure of nad+-dependent dna ligase
|
|
|
| 84 | c1q69B_ |
|
not modelled |
27.6 |
30 |
PDB header:membrane protein/transferase
Chain: B: PDB Molecule:proto-oncogene tyrosine-protein kinase lck;
PDBTitle: solution structure of t-cell surface glycoprotein cd8 alpha2 chain and proto-oncogene tyrosine-protein kinase lck3 fragments
|
|
|
| 85 | c1bi6H_ |
|
not modelled |
27.6 |
26 |
PDB header:cysteine protease inhibitor
Chain: H: PDB Molecule:bromelain inhibitor vi;
PDBTitle: nmr structure of bromelain inhibitor vi from pineapple stem
|
|
|
| 86 | c2bi6H_ |
|
not modelled |
27.6 |
26 |
PDB header:cysteine protease inhibitor
Chain: H: PDB Molecule:bromelain inhibitor vi;
PDBTitle: nmr study of bromelain inhibitor vi from pineapple stem
|
|
|
| 87 | c5y06A_ |
|
not modelled |
27.5 |
27 |
PDB header:unknown function
Chain: A: PDB Molecule:msmeg_4306;
PDBTitle: structural characterization of msmeg_4306 from mycobacterium smegmatis
|
|
|
| 88 | c3imkA_ |
|
not modelled |
26.9 |
19 |
PDB header:metal binding protein
Chain: A: PDB Molecule:putative molybdenum carrier protein;
PDBTitle: crystal structure of putative molybdenum carrier protein (yp_461806.1)2 from syntrophus aciditrophicus sb at 1.45 a resolution
|
|
|
| 89 | d1x62a1 |
|
not modelled |
26.8 |
27 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
|
|
| 90 | d1hk8a_ |
|
not modelled |
26.8 |
22 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:Class III anaerobic ribonucleotide reductase NRDD subunit |
|
|
| 91 | c1hk8A_ |
|
not modelled |
26.8 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:anaerobic ribonucleotide-triphosphate reductase;
PDBTitle: structural basis for allosteric substrate specificity regulation in2 class iii ribonucleotide reductases: nrdd in complex with dgtp
|
|
|
| 92 | c5kl1B_ |
|
not modelled |
26.7 |
21 |
PDB header:rna binding protein/rna
Chain: B: PDB Molecule:protein nanos;
PDBTitle: crystal structure of the pumilio-nos-hunchback rna complex
|
|
|
| 93 | c3bjiA_ |
|
not modelled |
26.3 |
26 |
PDB header:signaling protein
Chain: A: PDB Molecule:proto-oncogene vav;
PDBTitle: structural basis of promiscuous guanine nucleotide exchange2 by the t-cell essential vav1
|
|
|
| 94 | c5ijlA_ |
|
not modelled |
26.2 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:dna polymerase ii large subunit;
PDBTitle: d-family dna polymerase - dp2 subunit (catalytic subunit)
|
|
|
| 95 | c2f9iD_ |
|
not modelled |
25.5 |
11 |
PDB header:transferase
Chain: D: PDB Molecule:acetyl-coenzyme a carboxylase carboxyl transferase subunit
PDBTitle: crystal structure of the carboxyltransferase subunit of acc from2 staphylococcus aureus
|
|
|
| 96 | d2vnud2 |
|
not modelled |
25.0 |
21 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
|
|
| 97 | c1dvbA_ |
|
not modelled |
24.8 |
27 |
PDB header:electron transport
Chain: A: PDB Molecule:rubrerythrin;
PDBTitle: rubrerythrin
|
|
|
| 98 | c5xodB_ |
|
not modelled |
24.6 |
35 |
PDB header:transcription
Chain: B: PDB Molecule:ski oncogene;
PDBTitle: crystal structure of human smad2-ski complex
|
|
|
| 99 | d1eb7a1 |
|
not modelled |
24.4 |
17 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Di-heme cytochrome c peroxidase |
|
|
| 100 | c2kc1A_ |
|
not modelled |
24.2 |
22 |
PDB header:structural protein
Chain: A: PDB Molecule:mkiaa1027 protein;
PDBTitle: nmr structure of the f0 domain (residues 0-85) of the talin2 ferm domain
|
|
|
| 101 | c2lvuA_ |
|
not modelled |
24.1 |
40 |
PDB header:transcription
Chain: A: PDB Molecule:zinc finger and btb domain-containing protein 17;
PDBTitle: solution structure of miz-1 zinc finger 10
|
|
|
| 102 | d1nvmb2 |
|
not modelled |
23.8 |
28 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
|
|
| 103 | c4glxA_ |
|
not modelled |
23.3 |
16 |
PDB header:ligase/ligase inhibitor/dna
Chain: A: PDB Molecule:dna ligase;
PDBTitle: dna ligase a in complex with inhibitor
|
|
|
| 104 | d1nmla1 |
|
not modelled |
23.3 |
17 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Di-heme cytochrome c peroxidase |
|
|
| 105 | d6rxna_ |
|
not modelled |
23.1 |
30 |
Fold:Rubredoxin-like
Superfamily:Rubredoxin-like
Family:Rubredoxin |
|
|
| 106 | d1iqca1 |
|
not modelled |
22.8 |
17 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Di-heme cytochrome c peroxidase |
|
|
| 107 | d2coba1 |
|
not modelled |
22.7 |
15 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Psq domain |
|
|
| 108 | d1jk8a2 |
|
not modelled |
22.4 |
40 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
|
|
| 109 | c4jn6B_ |
|
not modelled |
22.2 |
34 |
PDB header:lyase/oxidoreductase
Chain: B: PDB Molecule:acetaldehyde dehydrogenase;
PDBTitle: crystal structure of the aldolase-dehydrogenase complex from2 mycobacterium tuberculosis hrv37
|
|
|
| 110 | c3uk2B_ |
|
not modelled |
21.9 |
24 |
PDB header:ligase
Chain: B: PDB Molecule:pantothenate synthetase;
PDBTitle: the structure of pantothenate synthetase from burkholderia2 thailandensis
|
|
|
| 111 | c3j3v0_ |
|
not modelled |
21.9 |
44 |
PDB header:ribosome
Chain: 0: PDB Molecule:50s ribosomal protein l32;
PDBTitle: atomic model of the immature 50s subunit from bacillus subtilis (state2 i-a)
|
|
|
| 112 | c2j04C_ |
|
not modelled |
21.7 |
25 |
PDB header:transcription
Chain: C: PDB Molecule:hypothetical protein ypl007c;
PDBTitle: the tau60-tau91 subcomplex of yeast transcription factor iiic
|
|
|
| 113 | c2f9yB_ |
|
not modelled |
21.7 |
28 |
PDB header:ligase
Chain: B: PDB Molecule:acetyl-coenzyme a carboxylase carboxyl transferase subunit
PDBTitle: the crystal structure of the carboxyltransferase subunit of acc from2 escherichia coli
|
|
|
| 114 | d2f9yb1 |
|
not modelled |
21.7 |
28 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
|
|
| 115 | c1pqrA_ |
|
not modelled |
21.7 |
43 |
PDB header:toxin
Chain: A: PDB Molecule:alpha-a-conotoxin eiva;
PDBTitle: solution conformation of alphaa-conotoxin eiva
|
|
|
| 116 | c5fmfR_ |
|
not modelled |
21.6 |
27 |
PDB header:transcription
Chain: R: PDB Molecule:transcription initiation factor iie subunit alpha, tfa1;
PDBTitle: the p-lobe of rna polymerase ii pre-initiation complex
|
|
|
| 117 | c5f4hF_ |
|
not modelled |
21.2 |
15 |
PDB header:hydrolase
Chain: F: PDB Molecule:nucleotide binding protein pinc;
PDBTitle: archael ruvb-like holiday junction helicase
|
|
|
| 118 | d1es0a2 |
|
not modelled |
21.2 |
40 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
|
|
| 119 | d1muja2 |
|
not modelled |
21.1 |
40 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
|
|
| 120 | c2fugC_ |
|
not modelled |
21.1 |
9 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh-quinone oxidoreductase chain 3;
PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
|
|
|