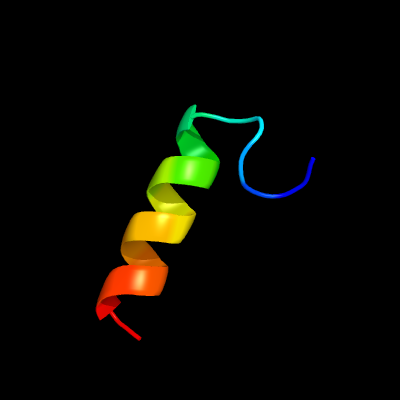
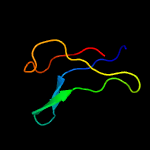
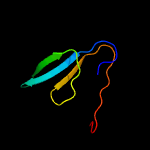
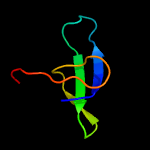
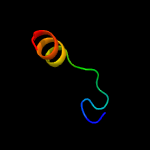
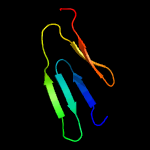
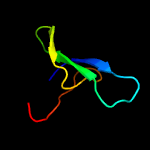
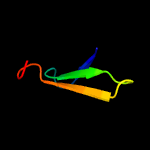
1 d1ppjw_
33.5
35
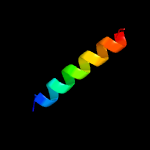
Fold: Single transmembrane helixSuperfamily: Subunit X (non-heme 7 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Family: Subunit X (non-heme 7 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
2 c3sulA_
29.8
33
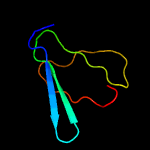
PDB header: unknown functionChain: A: PDB Molecule: cerato-platanin-like protein;PDBTitle: crystal structure of cerato-platanin 3 from m. perniciosa (mpcp3)
3 c5hceD_
24.5
24
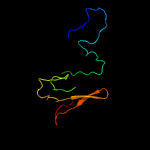
PDB header: immune systemChain: D: PDB Molecule: rhipicephalus appendiculatus raci1;PDBTitle: ternary complex of human complement c5 with ornithodoros moubata omci2 and rhipicephalus appendiculatus raci1
4 c3d0jA_
24.1
15
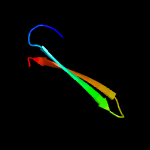
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ca_c3497;PDBTitle: crystal structure of conserved protein of unknown function ca_c34972 from clostridium acetobutylicum atcc 824
5 d2pi2e1
16.6
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB
6 d3cx5i1
16.2
25
Fold: Single transmembrane helixSuperfamily: Subunit X (non-heme 7 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Family: Subunit X (non-heme 7 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
7 d1vqoe1
16.1
15
Fold: Ribosomal protein L6Superfamily: Ribosomal protein L6Family: Ribosomal protein L6
8 c2pqaB_
15.1
18
PDB header: replicationChain: B: PDB Molecule: replication protein a 14 kda subunit;PDBTitle: crystal structure of full-length human rpa 14/32 heterodimer
9 c6az5A_
14.8
39
PDB header: sugar binding proteinChain: A: PDB Molecule: alpha-amylase;PDBTitle: crystal structure of cbmd (family cbm41) from eubacterium rectale2 amy13k
10 c5muzA_
14.0
24
PDB header: viral proteinChain: A: PDB Molecule: l protein;PDBTitle: structure of a c-terminal domain of a reptarenavirus l protein
11 c3k8rA_
13.8
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of protein of unknown function (yp_427503.1) from2 rhodospirillum rubrum atcc 11170 at 2.75 a resolution
12 c2fynO_
13.3
29
PDB header: oxidoreductaseChain: O: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfurPDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
13 c5e50A_
13.0
16
PDB header: lyaseChain: A: PDB Molecule: aprataxin and pnk-like factor;PDBTitle: aplf/xrcc4 complex
14 c2kklA_
12.7
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein mb1858;PDBTitle: solution nmr structure of fha domain of mb1858 from2 mycobacterium bovis. northeast structural genomics3 consortium target mbr243c (24-155).
15 c6f0kA_
12.4
14
PDB header: membrane proteinChain: A: PDB Molecule: cytochrome c family protein;PDBTitle: alternative complex iii
16 c5avoA_
12.4
22
PDB header: oxidoreductaseChain: A: PDB Molecule: homoserine dehydrogenase;PDBTitle: crystal structure of the reduced form of homoserine dehydrogenase from2 sulfolobus tokodaii.
17 c3sukB_
12.3
31
PDB header: unknown functionChain: B: PDB Molecule: cerato-platanin-like protein;PDBTitle: crystal structure of cerato-platanin 2 from m. perniciosa (mpcp2)
18 d1rl6a1
11.6
18
Fold: Ribosomal protein L6Superfamily: Ribosomal protein L6Family: Ribosomal protein L6
19 c3sujA_
10.2
27
PDB header: unknown functionChain: A: PDB Molecule: cerato-platanin 1;PDBTitle: crystal structure of cerato-platanin 1 from m. perniciosa (mpcp1)
20 c2n12A_
10.1
38
PDB header: motor protein,protein transportChain: A: PDB Molecule: unconventional myosin-vi;PDBTitle: solution structure of human myosin vi isoform3 (1050-1131)
21 c3sluB_
not modelled
9.9
16
PDB header: hydrolaseChain: B: PDB Molecule: m23 peptidase domain protein;PDBTitle: crystal structure of nmb0315
22 c3j21F_
not modelled
9.4
13
PDB header: ribosomeChain: F: PDB Molecule: 50s ribosomal protein l6p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
23 c3qr8A_
not modelled
8.7
15
PDB header: viral proteinChain: A: PDB Molecule: baseplate assembly protein v;PDBTitle: crystal structure of the bacteriophage p2 membrane-piercing protein2 gpv
24 d1zata2
not modelled
7.5
9
Fold: L,D-transpeptidase pre-catalytic domain-likeSuperfamily: L,D-transpeptidase pre-catalytic domain-likeFamily: L,D-transpeptidase pre-catalytic domain-like
25 c6hwhB_
not modelled
7.0
12
PDB header: electron transportChain: B: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfur subunit;PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
26 c3ljyA_
not modelled
6.7
16
PDB header: cell adhesionChain: A: PDB Molecule: putative adhesin;PDBTitle: crystal structure of putative adhesin (yp_001304413.1) from2 parabacteroides distasonis atcc 8503 at 2.41 a resolution
27 c3oggA_
not modelled
6.6
13
PDB header: toxinChain: A: PDB Molecule: botulinum neurotoxin type d;PDBTitle: crystal structure of the receptor binding domain of botulinum2 neurotoxin d
28 c2lowA_
not modelled
6.2
25
PDB header: membrane proteinChain: A: PDB Molecule: apelin receptor;PDBTitle: solution structure of ar55 in 50% hfip
29 c2kdcC_
not modelled
6.1
17
PDB header: transferaseChain: C: PDB Molecule: diacylglycerol kinase;PDBTitle: nmr solution structure of e. coli diacylglycerol kinase2 (dagk) in dpc micelles
30 c4tmdA_
not modelled
5.9
19
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: x-ray structure of putative uncharacterized protein (rv0999 ortholog)2 from mycobacterium smegmatis
31 c1vw4H_
not modelled
5.9
14
PDB header: ribosomeChain: H: PDB Molecule: 54s ribosomal protein l23, mitochondrial;PDBTitle: structure of the yeast mitochondrial large ribosomal subunit
32 c3j3wJ_
not modelled
5.9
17
PDB header: ribosomeChain: J: PDB Molecule: 50s ribosomal protein l13;PDBTitle: atomic model of the immature 50s subunit from bacillus subtilis (state2 ii-a)
33 d1k4za_
not modelled
5.8
18
Fold: Single-stranded right-handed beta-helixSuperfamily: C-terminal domain of adenylylcyclase associated proteinFamily: C-terminal domain of adenylylcyclase associated protein
34 d1q8da_
not modelled
5.7
38
Fold: GDNF receptor-likeSuperfamily: GDNF receptor-likeFamily: GDNF receptor-like
35 d1fcda3
not modelled
5.7
17
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domainFamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domain
36 c1yn3A_
not modelled
5.6
41
PDB header: toxin, protein bindingChain: A: PDB Molecule: truncated cell surface protein map-w;PDBTitle: crystal structures of eap domains from staphylococcus2 aureus reveal an unexpected homology to bacterial3 superantigens
37 d1l5pa_
not modelled
5.6
23
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related
38 c3petA_
not modelled
5.5
15
PDB header: cell adhesionChain: A: PDB Molecule: putative adhesin;PDBTitle: crystal structure of a putative adhesin (bf0245) from bacteroides2 fragilis nctc 9343 at 2.07 a resolution
39 c2m5sA_
not modelled
5.4
22
PDB header: viral proteinChain: A: PDB Molecule: coat protein;PDBTitle: high-resolution nmr structure and cryo-em imaging support multiple2 functional roles for the accessory i-domain of phage p22 coat protein
40 c1yn4A_
not modelled
5.3
29
PDB header: unknown functionChain: A: PDB Molecule: eaph1;PDBTitle: crystal structures of eap domains from staphylococcus2 aureus reveal an unexpected homology to bacterial3 superantigens
41 c1yn5B_
not modelled
5.3
29
PDB header: unknown functionChain: B: PDB Molecule: eaph2;PDBTitle: crystal structures of eap domains from staphylococcus2 aureus reveal an unexpected homology to bacterial3 superantigens