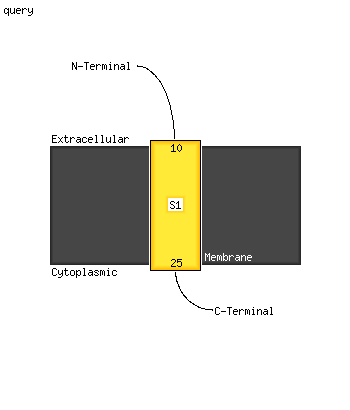
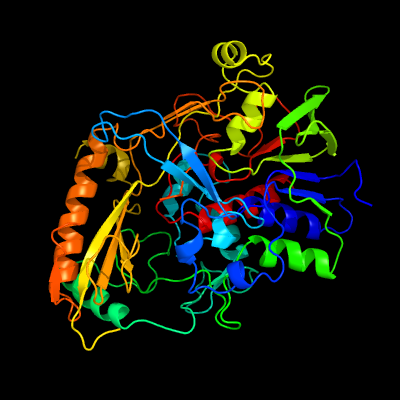
| 1 | c5hsaG_
|
|
 |
100.0 |
26 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:alcohol oxidase 1;
PDBTitle: alcohol oxidase aox1 from pichia pastoris
|
|
|
|
| 2 | c6h3gC_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:alcohol oxidase;
PDBTitle: alcohol oxidase from phanerochaete chrysosporium
|
|
|
|
| 3 | c5nccB_
|
|
 |
100.0 |
31 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:fatty acid photodecarboxylase;
PDBTitle: structure of fatty acid photodecarboxylase in complex with fad and2 palmitic acid
|
|
|
|
| 4 | c2jbvA_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:choline oxidase;
PDBTitle: crystal structure of choline oxidase reveals insights into the2 catalytic mechanism
|
|
|
|
| 5 | c3fimB_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aryl-alcohol oxidase;
PDBTitle: crystal structure of aryl-alcohol-oxidase from pleurotus eryingii
|
|
|
|
| 6 | c4yntA_
|
|
 |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucose oxidase, putative;
PDBTitle: crystal structure of aspergillus flavus fad glucose dehydrogenase
|
|
|
|
| 7 | c3t37A_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable dehydrogenase;
PDBTitle: crystal structure of pyridoxine 4-oxidase from mesorbium loti
|
|
|
|
| 8 | c1cf3A_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase(flavoprotein)
Chain: A: PDB Molecule:protein (glucose oxidase);
PDBTitle: glucose oxidase from apergillus niger
|
|
|
|
| 9 | c4h7uA_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyranose dehydrogenase;
PDBTitle: crystal structure of pyranose dehydrogenase from agaricus meleagris,2 wildtype
|
|
|
|
| 10 | c1gpeA_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase(flavoprotein)
Chain: A: PDB Molecule:protein (glucose oxidase);
PDBTitle: glucose oxidase from penicillium amagasakiense
|
|
|
|
| 11 | c4qi7A_
|
|
 |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cellobiose dehydrogenase;
PDBTitle: cellobiose dehydrogenase from neurospora crassa, nccdh
|
|
|
|
| 12 | c4qi6A_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cellobiose dehydrogenase;
PDBTitle: cellobiose dehydrogenase from myriococcum thermophilum, mtcdh
|
|
|
|
| 13 | c3q9tB_
|
|
 |
100.0 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:choline dehydrogenase and related flavoproteins;
PDBTitle: crystal structure analysis of formate oxidase
|
|
|
|
| 14 | c4qi4A_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cellobiose dehydrogenase;
PDBTitle: dehydrogenase domain of myriococcum thermophilum cellobiose2 dehydrogenase, mtdh
|
|
|
|
| 15 | c4udpA_
|
|
 |
100.0 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucose-methanol-choline oxidoreductase;
PDBTitle: crystal structure of 5-hydroxymethylfurfural oxidase (hmfo) in the2 oxidized state
|
|
|
|
| 16 | c1naaB_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cellobiose dehydrogenase;
PDBTitle: cellobiose dehydrogenase flavoprotein fragment in complex with2 cellobionolactam
|
|
|
|
| 17 | c1ju2A_
|
|
 |
100.0 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxynitrile lyase;
PDBTitle: crystal structure of the hydroxynitrile lyase from almond
|
|
|
|
| 18 | c4z24A_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:gmc-type oxidoreductase r135;
PDBTitle: mimivirus r135 (residues 51-702)
|
|
|
|
| 19 | c2gewA_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cholesterol oxidase;
PDBTitle: atomic resolution structure of cholesterol oxidase @ ph 9.02 (streptomyces sp. sa-coo)
|
|
|
|
| 20 | c1coyA_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase(oxygen receptor)
Chain: A: PDB Molecule:cholesterol oxidase;
PDBTitle: crystal structure of cholesterol oxidase complexed with a steroid2 substrate. implications for fad dependent alcohol oxidases
|
|
|
|
| 21 | c6a2uD_ |
|
not modelled |
100.0 |
19 |
PDB header:signaling protein/oxidoreductase
Chain: D: PDB Molecule:glucose dehydrogenase;
PDBTitle: crystal structure of gamma-alpha subunit complex from burkholderia2 cepacia fad glucose dehydrogenase
|
|
|
| 22 | c4migC_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:pyranose 2-oxidase;
PDBTitle: pyranose 2-oxidase from phanerochaete chrysosporium, recombinant wild2 type
|
|
|
| 23 | d1cf3a1 |
|
not modelled |
100.0 |
30 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 24 | c2igoG_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:pyranose oxidase;
PDBTitle: crystal structure of pyranose 2-oxidase h167a mutant with 2-fluoro-2-2 deoxy-d-glucose
|
|
|
| 25 | c2f5vA_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyranose 2-oxidase;
PDBTitle: reaction geometry and thermostability mutant of pyranose 2-oxidase2 from the white-rot fungus peniophora sp.
|
|
|
| 26 | d1kdga1 |
|
not modelled |
100.0 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 27 | d1gpea1 |
|
not modelled |
100.0 |
28 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 28 | d1n4wa1 |
|
not modelled |
100.0 |
25 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 29 | d3coxa1 |
|
not modelled |
100.0 |
24 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 30 | d2f5va1 |
|
not modelled |
100.0 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 31 | d1ju2a1 |
|
not modelled |
100.0 |
24 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 32 | c5glgA_ |
|
not modelled |
99.8 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fumarate reductase 2;
PDBTitle: the novel function of osm1 under anaerobic condition in the er was2 revealed by crystal structure of osm1, a soluble fumarate reductase3 in yeast
|
|
|
| 33 | c4repA_ |
|
not modelled |
99.8 |
13 |
PDB header:oxidoreductase, flavoprotein
Chain: A: PDB Molecule:gamma-carotene desaturase;
PDBTitle: crystal structure of gamma-carotenoid desaturase
|
|
|
| 34 | c1yq4A_ |
|
not modelled |
99.7 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:succinate dehydrogenase flavoprotein subunit;
PDBTitle: avian respiratory complex ii with 3-nitropropionate and ubiquinone
|
|
|
| 35 | c4at2A_ |
|
not modelled |
99.7 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-ketosteroid-delta4-5alpha-dehydrogenase;
PDBTitle: the crystal structure of 3-ketosteroid-delta4-(5alpha)-2 dehydrogenase from rhodococcus jostii rha1 in complex3 with 4-androstene-3,17- dione
|
|
|
| 36 | c4c3yF_ |
|
not modelled |
99.7 |
15 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:3-ketosteroid dehydrogenase;
PDBTitle: crystal structure of 3-ketosteroid delta1-dehydrogenase from2 rhodococcus erythropolis sq1 in complex with 1,4-androstadiene-3,17-3 dione
|
|
|
| 37 | c5xmjE_ |
|
not modelled |
99.7 |
17 |
PDB header:electron transport
Chain: E: PDB Molecule:fumarate reductase flavoprotein subunit;
PDBTitle: crystal structure of quinol:fumarate reductase from desulfovibrio2 gigas
|
|
|
| 38 | c3p4rM_ |
|
not modelled |
99.6 |
17 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:fumarate reductase flavoprotein subunit;
PDBTitle: crystal structure of menaquinol:fumarate oxidoreductase in complex2 with glutarate
|
|
|
| 39 | c3vr8E_ |
|
not modelled |
99.6 |
21 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:flavoprotein subunit of complex ii;
PDBTitle: mitochondrial rhodoquinol-fumarate reductase from the parasitic2 nematode ascaris suum
|
|
|
| 40 | c2aczA_ |
|
not modelled |
99.6 |
24 |
PDB header:oxidoreductase/electron transport
Chain: A: PDB Molecule:succinate dehydrogenase flavoprotein subunit;
PDBTitle: complex ii (succinate dehydrogenase) from e. coli with atpenin a52 inhibitor co-crystallized at the ubiquinone binding site
|
|
|
| 41 | c1qo8A_ |
|
not modelled |
99.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:flavocytochrome c3 fumarate reductase;
PDBTitle: the structure of the open conformation of a flavocytochrome c32 fumarate reductase
|
|
|
| 42 | c1d4cB_ |
|
not modelled |
99.6 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:flavocytochrome c fumarate reductase;
PDBTitle: crystal structure of the uncomplexed form of the flavocytochrome c2 fumarate reductase of shewanella putrefaciens strain mr-1
|
|
|
| 43 | c1kf6A_ |
|
not modelled |
99.6 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fumarate reductase flavoprotein;
PDBTitle: e. coli quinol-fumarate reductase with bound inhibitor hqno
|
|
|
| 44 | c1jrxA_ |
|
not modelled |
99.6 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:flavocytochrome c;
PDBTitle: crystal structure of arg402ala mutant flavocytochrome c32 from shewanella frigidimarina
|
|
|
| 45 | d1d5ta1 |
|
not modelled |
99.6 |
15 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:GDI-like N domain |
|
|
| 46 | d1y0pa2 |
|
not modelled |
99.5 |
21 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
|
|
| 47 | c2bs3A_ |
|
not modelled |
99.5 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:quinol-fumarate reductase flavoprotein subunit a;
PDBTitle: glu c180 -> gln variant quinol:fumarate reductase from2 wolinella succinogenes
|
|
|
| 48 | c2rghA_ |
|
not modelled |
99.5 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alpha-glycerophosphate oxidase;
PDBTitle: structure of alpha-glycerophosphate oxidase from2 streptococcus sp.: a template for the mitochondrial alpha-3 glycerophosphate dehydrogenase
|
|
|
| 49 | c3cirM_ |
|
not modelled |
99.5 |
17 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:fumarate reductase flavoprotein subunit;
PDBTitle: e. coli quinol fumarate reductase frda t234a mutation
|
|
|
| 50 | c5hxwF_ |
|
not modelled |
99.5 |
14 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:l-amino acid deaminase;
PDBTitle: l-amino acid deaminase from proteus vulgaris
|
|
|
| 51 | c6n56A_ |
|
not modelled |
99.5 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fumarate reductase, flavo protein subunit;
PDBTitle: crystal structure of fumarate reductase, flavo protein subunit, from2 helicobacter pylori g27
|
|
|
| 52 | d2bcgg1 |
|
not modelled |
99.5 |
14 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:GDI-like N domain |
|
|
| 53 | d1qo8a2 |
|
not modelled |
99.5 |
17 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
|
|
| 54 | c2rgoA_ |
|
not modelled |
99.5 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alpha-glycerophosphate oxidase;
PDBTitle: structure of alpha-glycerophosphate oxidase from2 streptococcus sp.: a template for the mitochondrial alpha-3 glycerophosphate dehydrogenase
|
|
|
| 55 | c4p9sA_ |
|
not modelled |
99.4 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dimethylglycine dehydrogenase;
PDBTitle: crystal structure of the mature form of rat dmgdh
|
|
|
| 56 | d1kf6a2 |
|
not modelled |
99.4 |
14 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
|
|
| 57 | c2fjaC_ |
|
not modelled |
99.4 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:adenylylsulfate reductase, subunit a;
PDBTitle: adenosine 5'-phosphosulfate reductase in complex with substrate
|
|
|
| 58 | c3nlcA_ |
|
not modelled |
99.4 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein vp0956;
PDBTitle: crystal structure of the vp0956 protein from vibrio parahaemolyticus.2 northeast structural genomics consortium target vpr147
|
|
|
| 59 | c2r4jA_ |
|
not modelled |
99.4 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aerobic glycerol-3-phosphate dehydrogenase;
PDBTitle: crystal structure of escherichia coli semet substituted glycerol-3-2 phosphate dehydrogenase in complex with dhap
|
|
|
| 60 | d2gf3a1 |
|
not modelled |
99.4 |
21 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 61 | c1pj6A_ |
|
not modelled |
99.4 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n,n-dimethylglycine oxidase;
PDBTitle: crystal structure of dimethylglycine oxidase of arthrobacter2 globiformis in complex with folic acid
|
|
|
| 62 | c4xwzA_ |
|
not modelled |
99.4 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fructosyl amine:oxygen oxidoreductase;
PDBTitle: the crystal structure of fructosyl amine: oxygen oxidoreductase2 (amadoriase i) from aspergillus fumigatus in complex with the3 substrate fructosyl lysine
|
|
|
| 63 | d1pj5a2 |
|
not modelled |
99.4 |
24 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 64 | d2gqfa1 |
|
not modelled |
99.4 |
18 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:HI0933 N-terminal domain-like |
|
|
| 65 | c1zkqA_ |
|
not modelled |
99.4 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase 2, mitochondrial;
PDBTitle: crystal structure of mouse thioredoxin reductase type 2
|
|
|
| 66 | c3dgzA_ |
|
not modelled |
99.4 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase 2;
PDBTitle: crystal structure of mouse mitochondrial thioredoxin reductase, c-2 terminal 3-residue truncation
|
|
|
| 67 | c3ka7A_ |
|
not modelled |
99.4 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of an oxidoreductase from methanosarcina2 mazei. northeast structural genomics consortium target id3 mar208
|
|
|
| 68 | d2bs2a2 |
|
not modelled |
99.4 |
17 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
|
|
| 69 | c3axbA_ |
|
not modelled |
99.4 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: structure of a dye-linked l-proline dehydrogenase from the aerobic2 hyperthermophilic archaeon, aeropyrum pernix
|
|
|
| 70 | c4x9mA_ |
|
not modelled |
99.4 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-alpha-glycerophosphate oxidase;
PDBTitle: oxidized l-alpha-glycerophosphate oxidase from mycoplasma pneumoniae2 with fad bound
|
|
|
| 71 | d1neka2 |
|
not modelled |
99.4 |
21 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
|
|
| 72 | c3da1A_ |
|
not modelled |
99.4 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol-3-phosphate dehydrogenase;
PDBTitle: x-ray structure of the glycerol-3-phosphate dehydrogenase2 from bacillus halodurans complexed with fad. northeast3 structural genomics consortium target bhr167.
|
|
|
| 73 | d1d4ca2 |
|
not modelled |
99.3 |
19 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
|
|
| 74 | c6c87A_ |
|
not modelled |
99.3 |
14 |
PDB header:protein transport
Chain: A: PDB Molecule:rab gdp dissociation inhibitor alpha;
PDBTitle: crystal structure of rab gdp dissociation inhibitor alpha from2 naegleria fowleri
|
|
|
| 75 | c2c3dB_ |
|
not modelled |
99.3 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-oxopropyl-com reductase;
PDBTitle: 2.15 angstrom crystal structure of 2-ketopropyl coenzyme m2 oxidoreductase carboxylase with a coenzyme m disulfide3 bound at the active site
|
|
|
| 76 | c3dmeB_ |
|
not modelled |
99.3 |
21 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:conserved exported protein;
PDBTitle: crystal structure of conserved exported protein from bordetella2 pertussis. northeast structural genomics target ber141
|
|
|
| 77 | c1gndA_ |
|
not modelled |
99.3 |
11 |
PDB header:gtpase activation
Chain: A: PDB Molecule:guanine nucleotide dissociation inhibitor;
PDBTitle: guanine nucleotide dissociation inhibitor, alpha-isoform
|
|
|
| 78 | c5fjnB_ |
|
not modelled |
99.3 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:l-amino acid deaminase;
PDBTitle: structure of l-amino acid deaminase from proteus myxofaciens2 in complex with anthranilate
|
|
|
| 79 | c2zxiC_ |
|
not modelled |
99.3 |
23 |
PDB header:fad-binding protein
Chain: C: PDB Molecule:trna uridine 5-carboxymethylaminomethyl modification enzyme
PDBTitle: structure of aquifex aeolicus gida in the form ii crystal
|
|
|
| 80 | c1ltxR_ |
|
not modelled |
99.3 |
12 |
PDB header:transferase/protein binding
Chain: R: PDB Molecule:rab escort protein 1;
PDBTitle: structure of rab escort protein-1 in complex with rab geranylgeranyl2 transferase and isoprenoid
|
|
|
| 81 | c2nvkX_ |
|
not modelled |
99.3 |
16 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:thioredoxin reductase;
PDBTitle: crystal structure of thioredoxin reductase from drosophila2 melanogaster
|
|
|
| 82 | d1ryia1 |
|
not modelled |
99.3 |
13 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 83 | c2olnA_ |
|
not modelled |
99.3 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nikd protein;
PDBTitle: nikd, an unusual amino acid oxidase essential for2 nikkomycin biosynthesis: closed form at 1.15 a resolution
|
|
|
| 84 | c2gahB_ |
|
not modelled |
99.3 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:heterotetrameric sarcosine oxidase beta-subunit;
PDBTitle: heterotetrameric sarcosine: structure of a diflavin2 metaloenzyme at 1.85 a resolution
|
|
|
| 85 | d2i0za1 |
|
not modelled |
99.3 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:HI0933 N-terminal domain-like |
|
|
| 86 | c3jskN_ |
|
not modelled |
99.3 |
32 |
PDB header:biosynthetic protein
Chain: N: PDB Molecule:cypbp37 protein;
PDBTitle: thiazole synthase from neurospora crassa
|
|
|
| 87 | c3gyxA_ |
|
not modelled |
99.3 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:adenylylsulfate reductase;
PDBTitle: crystal structure of adenylylsulfate reductase from2 desulfovibrio gigas
|
|
|
| 88 | c3cpiH_ |
|
not modelled |
99.2 |
12 |
PDB header:protein transport
Chain: H: PDB Molecule:rab gdp-dissociation inhibitor;
PDBTitle: crystal structure of yeast rab-gdi
|
|
|
| 89 | c3djeA_ |
|
not modelled |
99.2 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fructosyl amine: oxygen oxidoreductase;
PDBTitle: crystal structure of the deglycating enzyme fructosamine2 oxidase from aspergillus fumigatus (amadoriase ii) in3 complex with fsa
|
|
|
| 90 | c3g05B_ |
|
not modelled |
99.2 |
19 |
PDB header:rna binding protein
Chain: B: PDB Molecule:trna uridine 5-carboxymethylaminomethyl modification enzyme
PDBTitle: crystal structure of n-terminal domain (2-550) of e.coli mnmg
|
|
|
| 91 | d1chua2 |
|
not modelled |
99.2 |
30 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain |
|
|
| 92 | c1zmcG_ |
|
not modelled |
99.2 |
19 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: crystal structure of human dihydrolipoamide dehydrogenase2 complexed to nad+
|
|
|
| 93 | c6b4oB_ |
|
not modelled |
99.1 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:glutathione reductase;
PDBTitle: 1.73 angstrom resolution crystal structure of glutathione reductase2 from enterococcus faecalis in complex with fad
|
|
|
| 94 | c3cesB_ |
|
not modelled |
99.1 |
20 |
PDB header:rna binding protein
Chain: B: PDB Molecule:trna uridine 5-carboxymethylaminomethyl modification enzyme
PDBTitle: crystal structure of e.coli mnmg (gida), a highly-conserved trna2 modifying enzyme
|
|
|
| 95 | c6aonB_ |
|
not modelled |
99.1 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: 1.72 angstrom resolution crystal structure of 2-oxoglutarate2 dehydrogenase complex subunit dihydrolipoamide dehydrogenase from3 bordetella pertussis in complex with fad
|
|
|
| 96 | c4dgkA_ |
|
not modelled |
99.1 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phytoene dehydrogenase;
PDBTitle: crystal structure of phytoene desaturase crti from pantoea ananatis
|
|
|
| 97 | c6bz0C_ |
|
not modelled |
99.1 |
23 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: 1.83 angstrom resolution crystal structure of dihydrolipoyl2 dehydrogenase from acinetobacter baumannii in complex with fad.
|
|
|
| 98 | c1hyuA_ |
|
not modelled |
99.1 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alkyl hydroperoxide reductase subunit f;
PDBTitle: crystal structure of intact ahpf
|
|
|
| 99 | c6gg2A_ |
|
not modelled |
99.1 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:amino acid oxidase fmpa;
PDBTitle: the structure of fsqb from aspergillus fumigatus, a flavoenzyme of the2 amine oxidase family
|
|
|
| 100 | c3urhB_ |
|
not modelled |
99.1 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: crystal structure of a dihydrolipoamide dehydrogenase from2 sinorhizobium meliloti 1021
|
|
|
| 101 | c1ojtA_ |
|
not modelled |
99.1 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:surface protein;
PDBTitle: structure of dihydrolipoamide dehydrogenase
|
|
|
| 102 | c3cp8C_ |
|
not modelled |
99.1 |
20 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:trna uridine 5-carboxymethylaminomethyl
PDBTitle: crystal structure of gida from chlorobium tepidum
|
|
|
| 103 | d1o5wa1 |
|
not modelled |
99.1 |
11 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 104 | d1rp0a1 |
|
not modelled |
99.1 |
28 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:Thi4-like |
|
|
| 105 | d1cf3a2 |
|
not modelled |
99.1 |
15 |
Fold:FAD-linked reductases, C-terminal domain
Superfamily:FAD-linked reductases, C-terminal domain
Family:GMC oxidoreductases |
|
|
| 106 | c2eq8E_ |
|
not modelled |
99.1 |
20 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:pyruvate dehydrogenase complex, dihydrolipoamide
PDBTitle: crystal structure of lipoamide dehydrogenase from thermus thermophilus2 hb8 with psbdp
|
|
|
| 107 | c6mp5B_ |
|
not modelled |
99.1 |
17 |
PDB header:membrane protein, oxidoreductase
Chain: B: PDB Molecule:sulfide:quinone oxidoreductase, mitochondrial;
PDBTitle: crystal structure of native human sulfide:quinone oxidoreductase
|
|
|
| 108 | c4rslA_ |
|
not modelled |
99.1 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fructosyl peptide oxidase;
PDBTitle: structure of fructosyl peptide oxidase from e. terrenum
|
|
|
| 109 | d2cula1 |
|
not modelled |
99.1 |
28 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:GidA-like |
|
|
| 110 | d1ju2a2 |
|
not modelled |
99.1 |
18 |
Fold:FAD-linked reductases, C-terminal domain
Superfamily:FAD-linked reductases, C-terminal domain
Family:GMC oxidoreductases |
|
|
| 111 | c3cp2A_ |
|
not modelled |
99.1 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trna uridine 5-carboxymethylaminomethyl
PDBTitle: crystal structure of gida from e. coli
|
|
|
| 112 | c1m6iA_ |
|
not modelled |
99.1 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:programmed cell death protein 8;
PDBTitle: crystal structure of apoptosis inducing factor (aif)
|
|
|
| 113 | c1chuA_ |
|
not modelled |
99.1 |
32 |
PDB header:flavoenzyme
Chain: A: PDB Molecule:protein (l-aspartate oxidase);
PDBTitle: structure of l-aspartate oxidase: implications for the2 succinate dehydrogenase/ fumarate reducatse family
|
|
|
| 114 | c2a8xA_ |
|
not modelled |
99.1 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrolipoyl dehydrogenase;
PDBTitle: crystal structure of lipoamide dehydrogenase from2 mycobacterium tuberculosis
|
|
|
| 115 | c4y4nE_ |
|
not modelled |
99.0 |
26 |
PDB header:biosynthetic protein
Chain: E: PDB Molecule:putative ribose 1,5-bisphosphate isomerase;
PDBTitle: thiazole synthase thi4 from methanococcus igneus
|
|
|
| 116 | c3v76A_ |
|
not modelled |
99.0 |
20 |
PDB header:flavoprotein
Chain: A: PDB Molecule:flavoprotein;
PDBTitle: the crystal structure of a flavoprotein from sinorhizobium meliloti
|
|
|
| 117 | c2e5vA_ |
|
not modelled |
99.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-aspartate oxidase;
PDBTitle: crystal structure of l-aspartate oxidase from2 hyperthermophilic archaeon sulfolobus tokodaii
|
|
|
| 118 | d1gpea2 |
|
not modelled |
99.0 |
17 |
Fold:FAD-linked reductases, C-terminal domain
Superfamily:FAD-linked reductases, C-terminal domain
Family:GMC oxidoreductases |
|
|
| 119 | c1y56B_ |
|
not modelled |
99.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:sarcosine oxidase;
PDBTitle: crystal structure of l-proline dehydrogenase from p.horikoshii
|
|
|
| 120 | c5g3sB_ |
|
not modelled |
99.0 |
9 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-tryptophan oxidase vioa;
PDBTitle: the structure of the l-tryptophan oxidase vioa from chromobacterium2 violaceum - samarium derivative
|
|
|