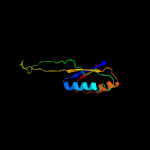
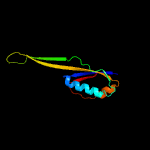
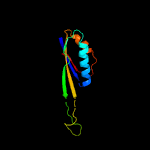
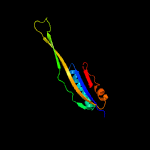
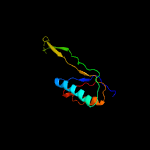
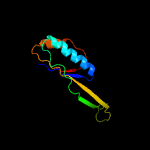
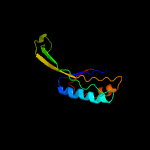
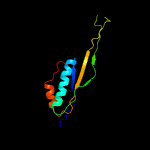
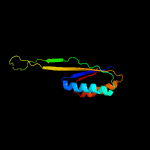
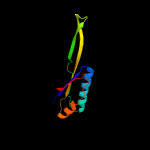
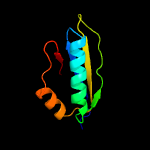
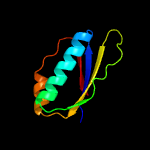
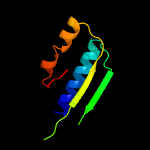
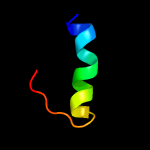
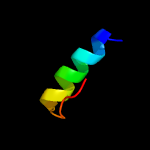
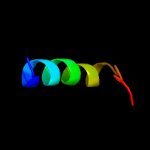1 c5o5jJ_
100.0
98
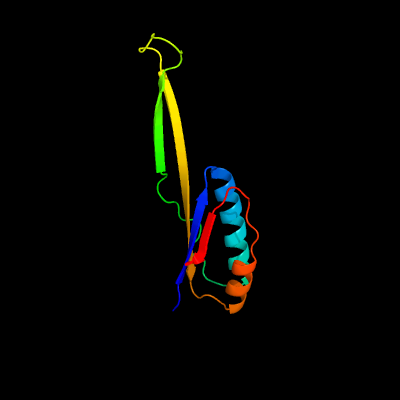
PDB header: ribosomeChain: J: PDB Molecule: 30s ribosomal protein s10;PDBTitle: structure of the 30s small ribosomal subunit from mycobacterium2 smegmatis
2 d2qalj1
100.0
61
Fold: Ferredoxin-likeSuperfamily: Ribosomal protein S10Family: Ribosomal protein S10
3 d2uubj1
100.0
58
Fold: Ferredoxin-likeSuperfamily: Ribosomal protein S10Family: Ribosomal protein S10
4 c3bbnJ_
100.0
53
PDB header: ribosomeChain: J: PDB Molecule: ribosomal protein s10;PDBTitle: homology model for the spinach chloroplast 30s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome.
5 c3zeyQ_
100.0
19
PDB header: ribosomeChain: Q: PDB Molecule: ribosomal protein s20, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
6 c3j20L_
100.0
35
PDB header: ribosomeChain: L: PDB Molecule: 30s ribosomal protein s10p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (30s3 ribosomal subunit)
7 c2xznJ_
100.0
27
PDB header: ribosomeChain: J: PDB Molecule: ribosomal protein s10 containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
8 c3j6vJ_
100.0
25
PDB header: ribosomeChain: J: PDB Molecule: 28s ribosomal protein s10, mitochondrial;PDBTitle: cryo-em structure of the small subunit of the mammalian mitochondrial2 ribosome
9 c2zkqj_
100.0
25
PDB header: ribosomal protein/rnaChain: J: PDB Molecule: PDBTitle: structure of a mammalian ribosomal 40s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
10 c1s1hJ_
100.0
29
PDB header: ribosomeChain: J: PDB Molecule: 40s ribosomal protein s20;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1h, contains 40s subunit. the 60s4 ribosomal subunit is in file 1s1i.
11 c3iz6J_
100.0
26
PDB header: ribosomeChain: J: PDB Molecule: 40s ribosomal protein s20 (s10p);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
12 c5xyiU_
100.0
24
PDB header: ribosomeChain: U: PDB Molecule: ribosomal protein s10p/s20e, putative;PDBTitle: small subunit of trichomonas vaginalis ribosome
13 c4v1ak_
99.9
20
PDB header: ribosomeChain: K: PDB Molecule: PDBTitle: structure of the large subunit of the mammalian mitoribosome, part 22 of 2
14 c2mewA_
99.9
70
PDB header: structural proteinChain: A: PDB Molecule: 30s ribosomal protein s10;PDBTitle: solution structure of nuse (s10) from thermotoga maritima
15 c3r2cJ_
99.9
57
PDB header: transcription/rnaChain: J: PDB Molecule: 30s ribosomal protein s10;PDBTitle: crystal structure of antitermination factors nusb and nuse in complex2 with boxa rna
16 c3j7yf_
99.3
17
PDB header: ribosomeChain: F: PDB Molecule: ul4;PDBTitle: structure of the large ribosomal subunit from human mitochondria
17 d1xbpg1
35.4
29
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Ribosomal protein L11, C-terminal domainFamily: Ribosomal protein L11, C-terminal domain
18 c3cjqB_
27.6
38
PDB header: transferase/ribosomal proteinChain: B: PDB Molecule: 50s ribosomal protein l11;PDBTitle: ribosomal protein l11 methyltransferase (prma) in complex with2 dimethylated ribosomal protein l11 in space group p212121
19 c4hubI_
21.1
24
PDB header: ribosomeChain: I: PDB Molecule: 50s ribosomal protein l11p;PDBTitle: the re-refined crystal structure of the haloarcula marismortui large2 ribosomal subunit at 2.4 angstrom resolution: more complete structure3 of the l7/l12 and l1 stalk, l5 and lx proteins
20 c5colB_
19.5
19
PDB header: translationChain: B: PDB Molecule: 50s ribosomal protein l11;PDBTitle: ribosomal protein l11 from methanococcus jannaschii
21 d1fnoa3
not modelled
18.7
21
Fold: Ferredoxin-likeSuperfamily: Bacterial exopeptidase dimerisation domainFamily: Bacterial exopeptidase dimerisation domain
22 d1hc8a_
not modelled
16.9
37
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Ribosomal protein L11, C-terminal domainFamily: Ribosomal protein L11, C-terminal domain
23 d1mmsa1
not modelled
16.8
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Ribosomal protein L11, C-terminal domainFamily: Ribosomal protein L11, C-terminal domain
24 c3ai4A_
not modelled
15.7
23
PDB header: fluorescent protein, replicationChain: A: PDB Molecule: yeast enhanced green fluorescent protein,dna polymerasePDBTitle: crystal structure of yeast enhanced green fluorescent protein - mouse2 polymerase iota ubiquitin binding motif fusion protein
25 c3j39K_
not modelled
15.5
19
PDB header: ribosomeChain: K: PDB Molecule: 60s ribosomal protein l12;PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
26 c1s1iK_
not modelled
14.9
21
PDB header: ribosomeChain: K: PDB Molecule: 60s ribosomal protein l12;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1i, contains 60s subunit. the 40s4 ribosomal subunit is in file 1s1h.
27 c2kl8A_
not modelled
13.8
14
PDB header: de novo proteinChain: A: PDB Molecule: or15;PDBTitle: solution nmr structure of de novo designed ferredoxin-like fold2 protein, northeast structural genomics consortium target or15
28 c3izcn_
not modelled
13.7
20
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein rpl14 (l14e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
29 c1vq8I_
not modelled
13.4
21
PDB header: ribosomeChain: I: PDB Molecule: 50s ribosomal protein l11p;PDBTitle: the structure of ccda-phe-cap-bio and the antibiotic sparsomycin bound2 to the large ribosomal subunit of haloarcula marismortui
30 c1s6xA_
not modelled
11.4
36
PDB header: toxinChain: A: PDB Molecule: kvap channel;PDBTitle: solution structure of vstx
31 c2zkri_
not modelled
11.1
21
PDB header: ribosomal protein/rnaChain: I: PDB Molecule: rna expansion segment es15 part i;PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
32 c2yqrA_
not modelled
11.0
24
PDB header: rna binding proteinChain: A: PDB Molecule: kiaa0907 protein;PDBTitle: solution structure of the kh domain in kiaa0907 protein
33 c5cw9A_
not modelled
10.9
14
PDB header: de novo proteinChain: A: PDB Molecule: de novo designed ferredoxin-ferredoxin domain insertionPDBTitle: crystal structure of de novo designed ferredoxin-ferredoxin domain2 insertion protein
34 d1dbda_
not modelled
10.9
19
Fold: Ferredoxin-likeSuperfamily: Viral DNA-binding domainFamily: Viral DNA-binding domain
35 d2bopa_
not modelled
10.6
19
Fold: Ferredoxin-likeSuperfamily: Viral DNA-binding domainFamily: Viral DNA-binding domain
36 c4ky3A_
not modelled
10.5
10
PDB header: de novo proteinChain: A: PDB Molecule: designed protein or327;PDBTitle: three-dimensional structure of the orthorhombic crystal of2 computationally designed insertion domain , northeast structural3 genomics consortium (nesg) target or327
37 d1iwga2
not modelled
10.4
15
Fold: Ferredoxin-likeSuperfamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomainsFamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains
38 c2ko4A_
not modelled
10.0
31
PDB header: transcriptionChain: A: PDB Molecule: mediator of rna polymerase ii transcription subunit 15;PDBTitle: complex structure of the activation domain of gcn4 bound to the2 mediator co-activator domain of gal11/med15
39 c1jqmA_
not modelled
9.8
26
PDB header: ribosomeChain: A: PDB Molecule: 50s ribosomal protein l11;PDBTitle: fitting of l11 protein and elongation factor g (ef-g) in2 the cryo-em map of e. coli 70s ribosome bound with ef-g,3 gdp and fusidic acid
40 c2k6nA_
not modelled
9.7
36
PDB header: structural proteinChain: A: PDB Molecule: supervillin;PDBTitle: solution structure of human supervillin headpiece, minimized2 average
41 c3j46n_
not modelled
9.6
33
PDB header: ribosome/protein transportChain: N: PDB Molecule: PDBTitle: structure of the secy protein translocation channel in action
42 c3zf7y_
not modelled
9.4
15
PDB header: ribosomeChain: Y: PDB Molecule: 60s ribosomal protein l24, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
43 c5o60J_
not modelled
9.4
42
PDB header: ribosomeChain: J: PDB Molecule: 50s ribosomal protein l11;PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
44 c3bboK_
not modelled
9.2
26
PDB header: ribosomeChain: K: PDB Molecule: ribosomal protein l11;PDBTitle: homology model for the spinach chloroplast 50s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome
45 c2vhmI_
not modelled
9.2
33
PDB header: ribosomeChain: I: PDB Molecule: 50s ribosomal protein l11;PDBTitle: structure of pdf binding helix in complex with the ribosome2 (part 1 of 4)
46 c4a1eE_
not modelled
9.2
11
PDB header: ribosomeChain: E: PDB Molecule: 60s ribosomal protein l9;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna, 5.8s rrna3 and proteins of molecule 1
47 c3lpeF_
not modelled
9.1
25
PDB header: transferaseChain: F: PDB Molecule: dna-directed rna polymerase subunit e'';PDBTitle: crystal structure of spt4/5ngn heterodimer complex from methanococcus2 jannaschii
48 c4p1zD_
not modelled
8.8
11
PDB header: rna binding proteinChain: D: PDB Molecule: piwi-like protein 1;PDBTitle: structure of the mid domain from miwi
49 d1vqoe2
not modelled
8.5
21
Fold: Ribosomal protein L6Superfamily: Ribosomal protein L6Family: Ribosomal protein L6
50 c4g0mB_
not modelled
8.5
11
PDB header: gene regulationChain: B: PDB Molecule: protein argonaute 2;PDBTitle: crystal structure of arabidopsis thaliana ago2 mid domain
51 c2kwvA_
not modelled
8.1
40
PDB header: protein binding/signaling proteinChain: A: PDB Molecule: dna polymerase iota;PDBTitle: solution structure of ubm1 of murine polymerase iota in complex with2 ubiquitin
52 d1yvua2
not modelled
8.1
5
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: PIWI domain
53 c3iz5F_
not modelled
7.9
22
PDB header: ribosomeChain: F: PDB Molecule: 60s ribosomal protein l9 (l6p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
54 c2hxgB_
not modelled
7.8
15
PDB header: isomeraseChain: B: PDB Molecule: l-arabinose isomerase;PDBTitle: crystal structure of mn2+ bound ecai
55 d1vqoi1
not modelled
7.6
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Ribosomal protein L11, C-terminal domainFamily: Ribosomal protein L11, C-terminal domain
56 c5h2wD_
not modelled
7.4
14
PDB header: protein transport/hydrolaseChain: D: PDB Molecule: ubiquitin-like-specific protease 1;PDBTitle: crystal structure of the karyopherin kap60p bound to the sumo protease2 ulp1p (150-340)
57 c5h2xB_
not modelled
7.4
14
PDB header: protein transport/hydrogenaseChain: B: PDB Molecule: ubiquitin-like-specific protease 1;PDBTitle: crystal structure of the karyopherin kap60p bound to the sumo protease2 ulp1p (150-172)
58 c3j39H_
not modelled
7.1
19
PDB header: ribosomeChain: H: PDB Molecule: 60s ribosomal protein l9;PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
59 c2ln3A_
not modelled
7.0
11
PDB header: de novo proteinChain: A: PDB Molecule: de novo designed protein or135;PDBTitle: solution nmr structure of de novo designed protein, if3-like fold,2 northeast structural genomics consortium target or135 (casd target)
60 c6amgA_
not modelled
6.7
8
PDB header: metal binding proteinChain: A: PDB Molecule: cytochrome p460;PDBTitle: cyt p460 of nitrosomonas sp. al212
61 d2q79a1
not modelled
6.6
12
Fold: Ferredoxin-likeSuperfamily: Viral DNA-binding domainFamily: Viral DNA-binding domain
62 c2mulA_
not modelled
6.6
50
PDB header: protein bindingChain: A: PDB Molecule: e3 ubiquitin-protein ligase huwe1;PDBTitle: solution structure of the ubm1 domain of human huwe1/arf-bp1
63 c3zf7M_
not modelled
6.3
16
PDB header: ribosomeChain: M: PDB Molecule: 60s ribosomal protein l12, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
64 c2l3xA_
not modelled
6.3
56
PDB header: protein bindingChain: A: PDB Molecule: ablim2 protein;PDBTitle: villin head piece domain of human ablim2
65 c3ccmE_
not modelled
6.3
19
PDB header: ribosomeChain: E: PDB Molecule: 50s ribosomal protein l6p;PDBTitle: structure of anisomycin resistant 50s ribosomal subunit: 23s rrna2 mutation g2611u
66 c3f41B_
not modelled
6.0
23
PDB header: hydrolaseChain: B: PDB Molecule: phytase;PDBTitle: structure of the tandemly repeated protein tyrosine2 phosphatase like phytase from mitsuokella multacida
67 c5an9B_
not modelled
5.9
19
PDB header: translationChain: B: PDB Molecule: 60s ribosomal protein l9;PDBTitle: mechanism of eif6 release from the nascent 60s ribosomal subunit
68 d1yu5x1
not modelled
5.8
71
Fold: VHP, Villin headpiece domainSuperfamily: VHP, Villin headpiece domainFamily: VHP, Villin headpiece domain
69 c3b0vD_
not modelled
5.8
15
PDB header: oxidoreductase/rnaChain: D: PDB Molecule: trna-dihydrouridine synthase;PDBTitle: trna-dihydrouridine synthase from thermus thermophilus in complex with2 trna
70 d1a7ge_
not modelled
5.5
15
Fold: Ferredoxin-likeSuperfamily: Viral DNA-binding domainFamily: Viral DNA-binding domain
71 d1j2jb_
not modelled
5.4
32
Fold: Spectrin repeat-likeSuperfamily: GAT-like domainFamily: GAT domain
72 c4bduC_
not modelled
5.4
24
PDB header: apoptosisChain: C: PDB Molecule: green fluorescent protein, apoptosis regulator bax;PDBTitle: bax bh3-in-groove dimer (gfp)
73 d1pbya3
not modelled
5.4
25
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Quinohemoprotein amine dehydrogenase A chain, domains 4 and 5
74 d1f9fa_
not modelled
5.3
11
Fold: Ferredoxin-likeSuperfamily: Viral DNA-binding domainFamily: Viral DNA-binding domain
75 c5o60W_
not modelled
5.2
20
PDB header: ribosomeChain: W: PDB Molecule: 50s ribosomal protein l25;PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
76 d1r8ha_
not modelled
5.2
4
Fold: Ferredoxin-likeSuperfamily: Viral DNA-binding domainFamily: Viral DNA-binding domain
77 d1qzpa_
not modelled
5.1
57
Fold: VHP, Villin headpiece domainSuperfamily: VHP, Villin headpiece domainFamily: VHP, Villin headpiece domain
78 c5vntA_
not modelled
5.1
29
PDB header: protein bindingChain: A: PDB Molecule: villin-4;PDBTitle: solution nmr structure of the c-terminal headpiece domain of villin 42 from a.thaliana, the first non-vertebrate headpiece structure