1 c5o60D_
100.0
88
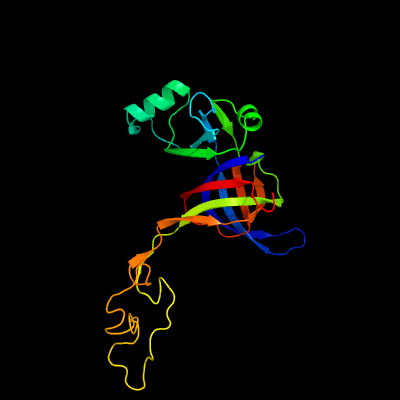
PDB header: ribosomeChain: D: PDB Molecule: 50s ribosomal protein l3;PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
2 c1vw3C_
100.0
41
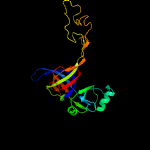
PDB header: ribosomeChain: C: PDB Molecule: 54s ribosomal protein l9, mitochondrial;PDBTitle: structure of the yeast mitochondrial large ribosomal subunit
3 c4v19E_
100.0
30
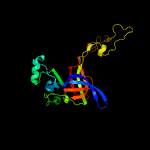
PDB header: ribosomeChain: E: PDB Molecule: mitoribosomal protein ul3m, mrpl3;PDBTitle: structure of the large subunit of the mammalian mitoribosome, part 12 of 2
4 d2gycb1
100.0
51
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Ribosomal protein L3
5 c3j3vD_
100.0
53
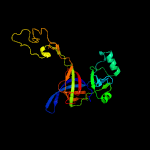
PDB header: ribosomeChain: D: PDB Molecule: 50s ribosomal protein l3;PDBTitle: atomic model of the immature 50s subunit from bacillus subtilis (state2 i-a)
6 c4wfbB_
100.0
52
PDB header: ribosomeChain: B: PDB Molecule: 50s ribosomal protein l3;PDBTitle: the crystal structure of the large ribosomal subunit of staphylococcus2 aureus in complex with bc-3205
7 d2zjrb1
100.0
53
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Ribosomal protein L3
8 c5mlcE_
100.0
43
PDB header: ribosomeChain: E: PDB Molecule: 50s ribosomal protein l3, chloroplastic;PDBTitle: cryo-em structure of the spinach chloroplast ribosome reveals the2 location of plastid-specific ribosomal proteins and extensions
9 c2ftcC_
100.0
33
PDB header: ribosomeChain: C: PDB Molecule: mitochondrial 39s ribosomal protein l3;PDBTitle: structural model for the large subunit of the mammalian mitochondrial2 ribosome
10 d2j01e1
100.0
56
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Ribosomal protein L3
11 d1vqob1
100.0
35
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Ribosomal protein L3
12 c3jywC_
100.0
27
PDB header: ribosomeChain: C: PDB Molecule: 60s ribosomal protein l3;PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
13 c3j21C_
100.0
30
PDB header: ribosomeChain: C: PDB Molecule: 50s ribosomal protein l3p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
14 c3u5iB_
100.0
27
PDB header: ribosomeChain: B: PDB Molecule: 60s ribosomal protein l3;PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution. this2 entry contains proteins of the 60s subunit, ribosome b
15 c1s1iC_
100.0
27
PDB header: ribosomeChain: C: PDB Molecule: 60s ribosomal protein l3;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1i, contains 60s subunit. the 40s4 ribosomal subunit is in file 1s1h.
16 c3iz5C_
100.0
26
PDB header: ribosomeChain: C: PDB Molecule: 60s ribosomal protein l3 (l3p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
17 c4a1aB_
100.0
27
PDB header: ribosomeChain: B: PDB Molecule: ribosomal protein l3;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
18 c3j39B_
100.0
24
PDB header: ribosomeChain: B: PDB Molecule: 60s ribosomal protein l3;PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
19 c2zkrb_
100.0
26
PDB header: ribosomal protein/rnaChain: B: PDB Molecule: rna expansion segment es4;PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
20 c3zf7f_
100.0
25
PDB header: ribosomeChain: F: PDB Molecule: PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
21 c5ancA_
not modelled
100.0
26
PDB header: translationChain: A: PDB Molecule: 60s ribosomal protein l3;PDBTitle: mechanism of eif6 release from the nascent 60s ribosomal subunit
22 c3bboF_
not modelled
100.0
42
PDB header: ribosomeChain: F: PDB Molecule: ribosomal protein l3;PDBTitle: homology model for the spinach chloroplast 50s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome
23 d1qfja1
not modelled
33.0
18
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like
24 c1a8pA_
not modelled
25.3
20
PDB header: oxidoreductaseChain: A: PDB Molecule: nadph\:ferredoxin oxidoreductase;PDBTitle: ferredoxin reductase from azotobacter vinelandii
25 c3j21R_
not modelled
23.6
33
PDB header: ribosomeChain: R: PDB Molecule: 50s ribosomal protein l21e;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
26 d1vqoq1
not modelled
22.7
44
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e
27 c1a3qA_
not modelled
20.0
15
PDB header: transcription/dnaChain: A: PDB Molecule: protein (nuclear factor kappa-b p52);PDBTitle: human nf-kappa-b p52 bound to dna
28 c4p6vF_
not modelled
19.6
28
PDB header: oxidoreductaseChain: F: PDB Molecule: na(+)-translocating nadh-quinone reductase subunit f;PDBTitle: crystal structure of the na+-translocating nadh: ubiquinone2 oxidoreductase from vibrio cholerae
29 c2zkrq_
not modelled
18.9
33
PDB header: ribosomal protein/rnaChain: Q: PDB Molecule: rna expansion segment es31 part ii;PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
30 d1fdra1
not modelled
18.2
20
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like
31 c2bruC_
not modelled
17.7
50
PDB header: oxidoreductaseChain: C: PDB Molecule: nad(p) transhydrogenase subunit beta;PDBTitle: complex of the domain i and domain iii of escherichia coli2 transhydrogenase
32 d1d4oa_
not modelled
16.7
50
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Transhydrogenase domain III (dIII)
33 c2l55A_
not modelled
16.3
8
PDB header: metal binding proteinChain: A: PDB Molecule: silb,silver efflux protein, mfp component of the threePDBTitle: solution structure of the c-terminal domain of silb from cupriavidus2 metallidurans
34 d1pnoa_
not modelled
15.9
67
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Transhydrogenase domain III (dIII)
35 c1pt9B_
not modelled
15.5
50
PDB header: oxidoreductaseChain: B: PDB Molecule: nad(p) transhydrogenase, mitochondrial;PDBTitle: crystal structure analysis of the diii component of transhydrogenase2 with a thio-nicotinamide nucleotide analogue
36 c3zf7U_
not modelled
13.8
67
PDB header: ribosomeChain: U: PDB Molecule: 60s ribosomal protein l21e, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
37 d1r4ya_
not modelled
13.8
23
Fold: Microbial ribonucleasesSuperfamily: Microbial ribonucleasesFamily: Ribotoxin
38 d1de3a_
not modelled
13.7
24
Fold: Microbial ribonucleasesSuperfamily: Microbial ribonucleasesFamily: Ribotoxin
39 c3j3bT_
not modelled
13.4
40
PDB header: ribosomeChain: T: PDB Molecule: 60s ribosomal protein l21;PDBTitle: structure of the human 60s ribosomal proteins
40 d1qcsa1
not modelled
13.3
19
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Cdc48 N-terminal domain-like
41 c1s1iQ_
not modelled
13.2
33
PDB header: ribosomeChain: Q: PDB Molecule: 60s ribosomal protein l21-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1i, contains 60s subunit. the 40s4 ribosomal subunit is in file 1s1h.
42 c5ogxA_
not modelled
13.0
21
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome p450 reductase;PDBTitle: crystal structure of amycolatopsis cytochrome p450 reductase gcob.
43 c3izcU_
not modelled
12.9
30
PDB header: ribosomeChain: U: PDB Molecule: 60s ribosomal protein rpl21 (l21e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
44 c4a1aP_
not modelled
12.8
40
PDB header: ribosomeChain: P: PDB Molecule: 60s ribosomal protein l21;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
45 c1le5E_
not modelled
12.5
26
PDB header: transcription/dnaChain: E: PDB Molecule: nuclear factor nf-kappa-b p65 subunit;PDBTitle: crystal structure of a nf-kb heterodimer bound to an ifnb-kb
46 d2i6va1
not modelled
12.4
20
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: EpsC C-terminal domain-like
47 c3iz5U_
not modelled
12.4
40
PDB header: ribosomeChain: U: PDB Molecule: 60s ribosomal protein l21 (l21e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
48 c4o9uB_
not modelled
12.3
67
PDB header: membrane proteinChain: B: PDB Molecule: nad(p) transhydrogenase subunit beta;PDBTitle: mechanism of transhydrogenase coupling proton translocation and2 hydride transfer
49 c4arpB_
not modelled
12.2
29
PDB header: hydrolaseChain: B: PDB Molecule: pesticin;PDBTitle: structure of the inactive pesticin e178a mutant
50 c3a98A_
not modelled
12.1
18
PDB header: signaling proteinChain: A: PDB Molecule: dedicator of cytokinesis protein 2;PDBTitle: crystal structure of the complex of the interacting regions of dock22 and elmo1
51 c2hdeA_
not modelled
12.1
40
PDB header: transcriptionChain: A: PDB Molecule: histone deacetylase complex subunit sap18;PDBTitle: solution structure of human sap18
52 c4bg7A_
not modelled
12.0
24
PDB header: replicationChain: A: PDB Molecule: putative transcriptional coactivator p15;PDBTitle: bacteriophage t5 homolog of the eukaryotic transcription coactivator2 pc4 implicated in recombination-dependent dna replication
53 c1qfjD_
not modelled
11.3
18
PDB header: oxidoreductaseChain: D: PDB Molecule: protein (flavin reductase);PDBTitle: crystal structure of nad(p)h:flavin oxidoreductase from escherichia2 coli
54 d2do3a1
not modelled
11.3
29
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: SPT5 KOW domain-like
55 d1a8pa1
not modelled
11.3
20
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like
56 c4wqmA_
not modelled
11.2
25
PDB header: oxidoreductaseChain: A: PDB Molecule: toluene-4-monooxygenase electron transfer component;PDBTitle: structure of the toluene 4-monooxygenase nadh oxidoreductase t4mof,2 k270s k271s variant
57 c6i52B_
not modelled
11.2
33
PDB header: dna binding proteinChain: B: PDB Molecule: replication factor a protein 2;PDBTitle: yeast rpa bound to ssdna
58 c4ytlB_
not modelled
11.0
33
PDB header: transcriptionChain: B: PDB Molecule: transcription elongation factor spt5;PDBTitle: structure of the kow2-kow3 domain of transcription elongation factor2 spt5.
59 c1nfiC_
not modelled
10.8
26
PDB header: complex (transcription reg/ank repeat)Chain: C: PDB Molecule: nf-kappa-b p65;PDBTitle: i-kappa-b-alpha/nf-kappa-b complex
60 c1zeqX_
not modelled
10.8
8
PDB header: metal binding proteinChain: X: PDB Molecule: cation efflux system protein cusf;PDBTitle: 1.5 a structure of apo-cusf residues 6-88 from escherichia2 coli
61 d1ikna2
not modelled
10.6
27
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: p53-like transcription factorsFamily: Rel/Dorsal transcription factors, DNA-binding domain
62 c1yb2A_
not modelled
10.4
0
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ta0852;PDBTitle: structure of a putative methyltransferase from thermoplasma2 acidophilum.
63 d1yb2a1
not modelled
10.4
0
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: tRNA(1-methyladenosine) methyltransferase-like
64 c5ccbB_
not modelled
10.4
31
PDB header: transferase/rnaChain: B: PDB Molecule: trna (adenine(58)-n(1))-methyltransferase non-catalyticPDBTitle: crystal structure of human m1a58 methyltransferase in a complex with2 trna3lys and sah
65 c1zuyB_
not modelled
10.3
12
PDB header: contractile proteinChain: B: PDB Molecule: myosin-5 isoform;PDBTitle: high-resolution structure of yeast myo5 sh3 domain
66 c5o60V_
not modelled
10.0
19
PDB header: ribosomeChain: V: PDB Molecule: 50s ribosomal protein l24;PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
67 d1uwfa1
not modelled
9.7
13
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits
68 c4b4dA_
not modelled
9.5
15
PDB header: oxidoreductaseChain: A: PDB Molecule: ferredoxin-nadp reductase;PDBTitle: crystal structure of fad-containing ferredoxin-nadp reductase from2 xanthomonas axonopodis pv. citri
69 d1vqot1
not modelled
9.4
25
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e
70 c4z89J_
not modelled
9.3
21
PDB header: rim-binding proteinChain: J: PDB Molecule: rim-binding protein, isoform f;PDBTitle: sh3-ii of drosophila rim-binding protein bound to a cacophony derived2 peptide
71 c1v1cA_
not modelled
8.9
35
PDB header: sh3-domainChain: A: PDB Molecule: obscurin;PDBTitle: solution structure of the sh3 domain of obscurin
72 c2kjpA_
not modelled
8.7
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ylbl;PDBTitle: solution structure of protein ylbl (bsu15050) from bacillus2 subtilis, northeast structural genomics consortium target3 sr713a
73 d1opka1
not modelled
8.6
12
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain
74 c5mmiV_
not modelled
8.6
25
PDB header: ribosomeChain: V: PDB Molecule: plastid ribosomal protein ul24c;PDBTitle: structure of the large subunit of the chloroplast ribosome
75 c5ergA_
not modelled
8.6
19
PDB header: transferaseChain: A: PDB Molecule: trna (adenine(58)-n(1))-methyltransferase non-catalyticPDBTitle: crystal structure of the two-subunit trna m1a58 methyltransferase2 trm6-trm61 in complex with sam
76 d2zjrr1
not modelled
8.5
13
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e
77 c3iz5Y_
not modelled
8.5
19
PDB header: ribosomeChain: Y: PDB Molecule: 60s ribosomal protein l26 (l24p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
78 c3j21U_
not modelled
8.2
25
PDB header: ribosomeChain: U: PDB Molecule: 50s ribosomal protein l24p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
79 d1vlra2
not modelled
8.2
16
Fold: mRNA decapping enzyme DcpS N-terminal domainSuperfamily: mRNA decapping enzyme DcpS N-terminal domainFamily: mRNA decapping enzyme DcpS N-terminal domain
80 c1cqxB_
not modelled
8.1
13
PDB header: lipid binding proteinChain: B: PDB Molecule: flavohemoprotein;PDBTitle: crystal structure of the flavohemoglobin from alcaligenes eutrophus at2 1.75 a resolution
81 c2i0nA_
not modelled
8.1
9
PDB header: structural proteinChain: A: PDB Molecule: class vii unconventional myosin;PDBTitle: structure of dictyostelium discoideum myosin vii sh3 domain2 with adjacent proline rich region
82 c4a1cS_
not modelled
8.0
13
PDB header: ribosomeChain: S: PDB Molecule: rpl26;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
83 c4tu3X_
not modelled
8.0
32
PDB header: hydrolase/protein transportChain: X: PDB Molecule: phosphoinositide phosphatase sac1;PDBTitle: crystal structure of yeast sac1/vps74 complex
84 c1qgyA_
not modelled
7.6
11
PDB header: oxidoreductaseChain: A: PDB Molecule: ferredoxin--nadp+ reductase;PDBTitle: ferredoxin:nadp+ reductase mutant with lys 75 replaced by glu (k75e)
85 c3iuwA_
not modelled
7.5
38
PDB header: rna binding proteinChain: A: PDB Molecule: activating signal cointegrator;PDBTitle: crystal structure of activating signal cointegrator (np_814290.1) from2 enterococcus faecalis v583 at 1.58 a resolution
86 c2lj0A_
not modelled
7.2
29
PDB header: signaling proteinChain: A: PDB Molecule: sorbin and sh3 domain-containing protein 1;PDBTitle: the third sh3 domain of r85fl
87 d1ra0a1
not modelled
7.1
26
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: Cytosine deaminase
88 c3x2dM_
not modelled
7.1
50
PDB header: viral protein/immune systemChain: M: PDB Molecule: envelope glycoprotein gp1;PDBTitle: crystal structure of marburg virus gp in complex with the human2 survivor antibody mr78
89 d1sm4a1
not modelled
6.9
20
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like
90 c1vpzB_
not modelled
6.9
36
PDB header: rna binding proteinChain: B: PDB Molecule: carbon storage regulator homolog;PDBTitle: crystal structure of a putative carbon storage regulator protein2 (csra, pa0905) from pseudomonas aeruginosa at 2.05 a resolution
91 c4pogC_
not modelled
6.8
33
PDB header: replication, dna binding protein/dnaChain: C: PDB Molecule: cell division control protein 21;PDBTitle: mcm-ssdna co-crystal structure
92 c5o0xA_
not modelled
6.8
18
PDB header: oxidoreductaseChain: A: PDB Molecule: putative ferric reductase;PDBTitle: crystal structure of dehydrogenase domain of cylindrospermum stagnale2 nadph-oxidase 5 (nox5)
93 c2rqrA_
not modelled
6.7
17
PDB header: protein bindingChain: A: PDB Molecule: engulfment and cell motility protein 1, linker, dedicatorPDBTitle: the solution structure of human dock2 sh3 domain - elmo1 peptide2 chimera complex
94 c4y6tD_
not modelled
6.7
23
PDB header: structural proteinChain: D: PDB Molecule: coat protein;PDBTitle: structure of tobacco streak virus coat protein dimer at 2.4 angstroms2 resolution
95 c3s88I_
not modelled
6.7
63
PDB header: immune system/viral proteinChain: I: PDB Molecule: envelope glycoprotein;PDBTitle: crystal structure of sudan ebolavirus glycoprotein (strain gulu) bound2 to 16f6
96 c1krhA_
not modelled
6.6
32
PDB header: oxidoreductaseChain: A: PDB Molecule: benzoate 1,2-dioxygenase reductase;PDBTitle: x-ray structure of benzoate dioxygenase reductase
97 c2vl6C_
not modelled
6.4
33
PDB header: dna binding proteinChain: C: PDB Molecule: minichromosome maintenance protein mcm;PDBTitle: structural analysis of the sulfolobus solfataricus mcm2 protein n-terminal domain
98 c4g1bB_
not modelled
6.3
30
PDB header: oxidoreductaseChain: B: PDB Molecule: flavohemoglobin;PDBTitle: x-ray structure of yeast flavohemoglobin in complex with econazole
99 c5j21C_
not modelled
6.2
20
PDB header: hydrolaseChain: C: PDB Molecule: bifunctional oligoribonuclease and pap phosphatase nrna;PDBTitle: structure of bacillus nanornase a (wt)