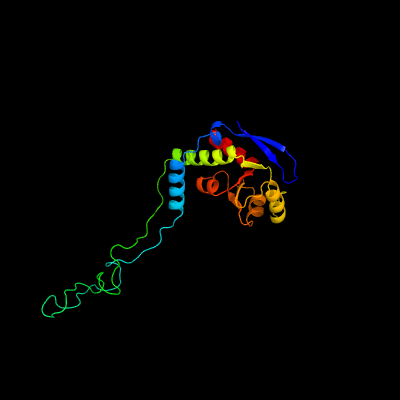
1 c5o60E_
100.0
87
PDB header: ribosomeChain: E: PDB Molecule: 50s ribosomal protein l4;PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
2 c4v19F_
100.0
28
PDB header: ribosomeChain: F: PDB Molecule: mitoribosomal protein ul4m, mrpl4;PDBTitle: structure of the large subunit of the mammalian mitoribosome, part 12 of 2
3 c3j3vE_
100.0
40
PDB header: ribosomeChain: E: PDB Molecule: 50s ribosomal protein l4;PDBTitle: atomic model of the immature 50s subunit from bacillus subtilis (state2 i-a)
4 c3bboG_
100.0
32
PDB header: ribosomeChain: G: PDB Molecule: ribosomal protein l4;PDBTitle: homology model for the spinach chloroplast 50s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome
5 c4wf9C_
100.0
43
PDB header: ribosomeChain: C: PDB Molecule: 50s ribosomal protein l4;PDBTitle: the crystal structure of the large ribosomal subunit of staphylococcus2 aureus in complex with telithromycin
6 c1vw3D_
100.0
23
PDB header: ribosomeChain: D: PDB Molecule: 54s ribosomal protein yml6, mitochondrial;PDBTitle: structure of the yeast mitochondrial large ribosomal subunit
7 c4ce4F_
100.0
28
PDB header: ribosomeChain: F: PDB Molecule: mrpl4;PDBTitle: 39s large subunit of the porcine mitochondrial ribosome
8 d2j01f1
100.0
38
Fold: Ribosomal protein L4Superfamily: Ribosomal protein L4Family: Ribosomal protein L4
9 d2gycc1
100.0
36
Fold: Ribosomal protein L4Superfamily: Ribosomal protein L4Family: Ribosomal protein L4
10 d2zjrc1
100.0
35
Fold: Ribosomal protein L4Superfamily: Ribosomal protein L4Family: Ribosomal protein L4
11 c3j21D_
100.0
26
PDB header: ribosomeChain: D: PDB Molecule: 50s ribosomal protein l4p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
12 c5tqbA_
100.0
18
PDB header: ribosomal proteinChain: A: PDB Molecule: 60s ribosomal protein l4-like protein;PDBTitle: crystal structure of assembly chaperone of ribosomal protein l4 (acl4)2 in complex with ribosomal protein l4 (rpl4)
13 c3jywD_
100.0
22
PDB header: ribosomeChain: D: PDB Molecule: 60s ribosomal protein l4(b);PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
14 d1vqoc1
100.0
23
Fold: Ribosomal protein L4Superfamily: Ribosomal protein L4Family: Ribosomal protein L4
15 c2zkrc_
100.0
24
PDB header: ribosomal protein/rnaChain: C: PDB Molecule: rna expansion segment es5;PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
16 c1s1iD_
100.0
22
PDB header: ribosomeChain: D: PDB Molecule: 60s ribosomal protein l4-b;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1i, contains 60s subunit. the 40s4 ribosomal subunit is in file 1s1h.
17 c3j39C_
100.0
27
PDB header: ribosomeChain: C: PDB Molecule: 60s ribosomal protein l4;PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
18 c4b6aC_
100.0
26
PDB header: ribosomeChain: C: PDB Molecule: 60s ribosomal protein l4-a;PDBTitle: cryo-em structure of the 60s ribosomal subunit in complex2 with arx1 and rei1
19 c2ftcD_
100.0
32
PDB header: ribosomeChain: D: PDB Molecule: mitochondrial ribosomal protein l4 isoform a;PDBTitle: structural model for the large subunit of the mammalian mitochondrial2 ribosome
20 c3iz5D_
100.0
28
PDB header: ribosomeChain: D: PDB Molecule: 60s ribosomal protein l4 (l4p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
21 c3j3bC_
not modelled
100.0
26
PDB header: ribosomeChain: C: PDB Molecule: 60s ribosomal protein l4;PDBTitle: structure of the human 60s ribosomal proteins
22 c3zf7r_
not modelled
100.0
21
PDB header: ribosomeChain: R: PDB Molecule: 60s ribosomal protein l17, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
23 c4a1aC_
not modelled
100.0
28
PDB header: ribosomeChain: C: PDB Molecule: rpl4;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
24 d1dmga_
not modelled
100.0
30
Fold: Ribosomal protein L4Superfamily: Ribosomal protein L4Family: Ribosomal protein L4
25 c2d3wB_
not modelled
37.6
8
PDB header: biosynthetic proteinChain: B: PDB Molecule: probable atp-dependent transporter sufc;PDBTitle: crystal structure of escherichia coli sufc, an atpase2 compenent of the suf iron-sulfur cluster assembly machinery
26 c1f2uD_
not modelled
21.9
9
PDB header: replicationChain: D: PDB Molecule: rad50 abc-atpase;PDBTitle: crystal structure of rad50 abc-atpase
27 c6bzrA_
not modelled
17.6
26
PDB header: transport proteinChain: A: PDB Molecule: multidrug resistance-associated protein 6;PDBTitle: human abcc6 nbd2 in adp-bound state
28 c2ehjA_
not modelled
17.6
13
PDB header: transferaseChain: A: PDB Molecule: uracil phosphoribosyltransferase;PDBTitle: structure of uracil phosphoribosyl transferase
29 d1fsga_
not modelled
17.1
19
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
30 c5xjyA_
not modelled
16.8
15
PDB header: transport proteinChain: A: PDB Molecule: atp-binding cassette sub-family a member 1;PDBTitle: cryo-em structure of human abca1
31 d2pmka1
not modelled
16.4
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like
32 c5a5uB_
not modelled
15.4
9
PDB header: translationChain: B: PDB Molecule: eukaryotic translation initiation factor 3 subunit b;PDBTitle: structure of mammalian eif3 in the context of the 43s preinitiation2 complex
33 c5kapA_
not modelled
15.4
24
PDB header: transferase/transferase inhibitorChain: A: PDB Molecule: hypoxanthine-guanine phosphoribosyltransferase;PDBTitle: trypanosome brucei hypoxanthine-guanine phosphoribosyltranferase in2 complex with a 9-(4-(phosphonobutil)hypoxanthine
34 d2gycj1
not modelled
15.0
39
Fold: Ribosomal proteins L15p and L18eSuperfamily: Ribosomal proteins L15p and L18eFamily: Ribosomal proteins L15p and L18e
35 c6n9lA_
not modelled
13.8
19
PDB header: dna binding proteinChain: A: PDB Molecule: uvrabc system protein a;PDBTitle: crystal structure of t. maritima uvra d117-399 with adp
36 d1cjba_
not modelled
13.2
19
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
37 c5t5dA_
not modelled
13.1
12
PDB header: transport proteinChain: A: PDB Molecule: pts system, iib component;PDBTitle: crystal structure of the pts iib protein associated with the fucose2 utilization operon from streptococcus pneumoniae
38 c1x37A_
not modelled
12.6
17
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent protease la 1;PDBTitle: structure of bacillus subtilis lon protease ssd domain
39 d1pf4a1
not modelled
12.3
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like
40 c1gpjA_
not modelled
12.3
16
PDB header: reductaseChain: A: PDB Molecule: glutamyl-trna reductase;PDBTitle: glutamyl-trna reductase from methanopyrus kandleri
41 c3nvbA_
not modelled
12.1
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of n-terminal part of the protein bf1531 from2 bacteroides fragilis containing phosphatase domain complexed with mg3 and tungstate
42 d1tc1a_
not modelled
12.1
22
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
43 c4gitA_
not modelled
12.0
25
PDB header: hydrolaseChain: A: PDB Molecule: lon protease;PDBTitle: crystal structure of alpha sub-domain of lon protease from2 brevibacillus thermoruber
44 c5xeiA_
not modelled
11.9
17
PDB header: dna binding protein, cell cycleChain: A: PDB Molecule: chromosome partition protein smc;PDBTitle: crystal structure of the smc head domain with a coiled coil and joint2 derived from pyrococcus yayanosii
45 c3ozxA_
not modelled
11.6
11
PDB header: hydrolase, translationChain: A: PDB Molecule: rnase l inhibitor;PDBTitle: crystal structure of abce1 of sulfolubus solfataricus (-fes domain)
46 c3h0gE_
not modelled
11.4
10
PDB header: transcriptionChain: E: PDB Molecule: dna-directed rna polymerases i, ii, and iii subunit rpabc1;PDBTitle: rna polymerase ii from schizosaccharomyces pombe
47 c5ipfA_
not modelled
11.3
22
PDB header: transferaseChain: A: PDB Molecule: hypoxanthine-guanine phosphoribosyltransferase (hgprt);PDBTitle: crystal structure of hypoxanthine-guanine phosphoribosyltransferase2 from schistosoma mansoni in complex with imp
48 c3zqjF_
not modelled
11.1
12
PDB header: dna binding proteinChain: F: PDB Molecule: uvrabc system protein a;PDBTitle: mycobacterium tuberculosis uvra
49 d1qzma_
not modelled
10.5
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
50 d2hyda1
not modelled
10.4
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like
51 c2wc1A_
not modelled
9.4
11
PDB header: electron transportChain: A: PDB Molecule: flavodoxin;PDBTitle: three-dimensional structure of the nitrogen fixation2 flavodoxin (niff) from rhodobacter capsulatus at 2.2 a
52 c4gu0F_
not modelled
9.4
35
PDB header: oxidoreductaseChain: F: PDB Molecule: histone h3.3;PDBTitle: crystal structure of lsd2 with h3
53 c4hsuC_
not modelled
9.4
35
PDB header: oxidoreductaseChain: C: PDB Molecule: histone h3;PDBTitle: crystal structure of lsd2-npac with h3(1-26)in space group p21
54 c4gu0E_
not modelled
9.4
35
PDB header: oxidoreductaseChain: E: PDB Molecule: histone h3.3;PDBTitle: crystal structure of lsd2 with h3
55 c3j3bq_
not modelled
9.2
14
PDB header: ribosomeChain: Q: PDB Molecule: 60s ribosomal protein l18;PDBTitle: structure of the human 60s ribosomal proteins
56 c4mrnB_
not modelled
8.5
13
PDB header: transport proteinChain: B: PDB Molecule: abc transporter related protein;PDBTitle: structure of a bacterial atm1-family abc transporter
57 c5idvA_
not modelled
8.5
13
PDB header: transport proteinChain: A: PDB Molecule: lipid a export atp-binding/permease protein msba;PDBTitle: structure of the nucleotide binding domain of an abc transporter msba2 from acinetobacter baumannii
58 d1mv5a_
not modelled
8.4
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like
59 c1xxiF_
not modelled
8.3
13
PDB header: transferaseChain: F: PDB Molecule: dna polymerase iii, delta subunit;PDBTitle: adp bound e. coli clamp loader complex
60 c2hydB_
not modelled
8.3
15
PDB header: transport proteinChain: B: PDB Molecule: abc transporter homolog;PDBTitle: multidrug abc transporter sav1866
61 c5tz8C_
not modelled
8.2
16
PDB header: transferaseChain: C: PDB Molecule: glycosyl transferase;PDBTitle: crystal structure of s. aureus tars
62 c3if4C_
not modelled
8.0
50
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: integron cassette protein hfx_cass5;PDBTitle: structure from the mobile metagenome of north west arm sewage outfall:2 integron cassette protein hfx_cass5
63 c5vykC_
not modelled
7.8
12
PDB header: signaling proteinChain: C: PDB Molecule: chimera protein of brs domain of braf and cc-sam domain ofPDBTitle: crystal structure of the brs domain of braf in complex with the cc-sam2 domain of ksr1
64 d3b60a1
not modelled
7.8
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like
65 d1z7ga1
not modelled
7.4
19
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
66 d1uhwa_
not modelled
7.1
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: DEP domain
67 c3qf4A_
not modelled
6.9
13
PDB header: transport proteinChain: A: PDB Molecule: abc transporter, atp-binding protein;PDBTitle: crystal structure of a heterodimeric abc transporter in its inward-2 facing conformation
68 c2kftB_
not modelled
6.8
37
PDB header: transcription/protein bindingChain: B: PDB Molecule: histone h3;PDBTitle: nmr solution structure of the first phd finger domain of2 human autoimmune regulator (aire) in complex with histone3 h3(1-20cys) peptide
69 c3iz5s_
not modelled
6.7
22
PDB header: ribosomeChain: S: PDB Molecule: 60s ribosomal protein l18a (l18ae);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
70 c3izcs_
not modelled
6.7
17
PDB header: ribosomeChain: S: PDB Molecule: 60s ribosomal protein rpl20 (l18ae);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
71 c2vf7B_
not modelled
6.5
17
PDB header: dna binding proteinChain: B: PDB Molecule: excinuclease abc, subunit a.;PDBTitle: crystal structure of uvra2 from deinococcus radiodurans
72 c3kb8A_
not modelled
6.3
26
PDB header: transferaseChain: A: PDB Molecule: hypoxanthine phosphoribosyltransferase;PDBTitle: 2.09 angstrom resolution structure of a hypoxanthine-guanine2 phosphoribosyltransferase (hpt-1) from bacillus anthracis str. 'ames3 ancestor' in complex with gmp
73 c2v3jA_
not modelled
6.1
18
PDB header: ribosomal proteinChain: A: PDB Molecule: essential for mitotic growth 1;PDBTitle: the yeast ribosome synthesis factor emg1 alpha beta knot2 fold methyltransferase
74 c4pl0B_
not modelled
6.0
11
PDB header: transport proteinChain: B: PDB Molecule: microcin-j25 export atp-binding/permease protein mcjd;PDBTitle: crystal structure of the antibacterial peptide abc transporter mcjd in2 an outward occluded state
75 d1oeyj_
not modelled
5.8
13
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: PB1 domain
76 c5ochF_
not modelled
5.8
17
PDB header: hydrolaseChain: F: PDB Molecule: atp-binding cassette sub-family b member 8, mitochondrial;PDBTitle: the crystal structure of human abcb8 in an outward-facing state
77 d1v2ya_
not modelled
5.8
12
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ubiquitin-related
78 c3htrB_
not modelled
5.7
43
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized prc-barrel domain protein;PDBTitle: crystal structure of prc-barrel domain protein from2 rhodopseudomonas palustris
79 c5e38D_
not modelled
5.6
10
PDB header: transferaseChain: D: PDB Molecule: uracil phosphoribosyltransferase;PDBTitle: structural basis of mapping the spontaneous mutations with 5-2 flourouracil in uracil phosphoribosyltransferase from mycobacterium3 tuberculosis
80 c5mvrA_
not modelled
5.5
15
PDB header: transferaseChain: A: PDB Molecule: trna threonylcarbamoyladenosine biosynthesis protein tsae;PDBTitle: crystal structure of bacillus subtilus ydib
81 d2v3ka1
not modelled
5.4
18
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: EMG1/NEP1-like
82 d1oxxk2
not modelled
5.4
25
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like
83 c4fwiB_
not modelled
5.3
13
PDB header: transport proteinChain: B: PDB Molecule: abc-type dipeptide/oligopeptide/nickel transport system,PDBTitle: crystal structure of the nucleotide-binding domain of a dipeptide abc2 transporter
84 c1pzmB_
not modelled
5.2
20
PDB header: transferaseChain: B: PDB Molecule: hypoxanthine-guanine phosphoribosyltransferase;PDBTitle: crystal structure of hgprt-ase from leishmania tarentolae in complex2 with gmp
85 c4u02C_
not modelled
5.2
14
PDB header: transport proteinChain: C: PDB Molecule: amino acid abc transporter, atp-binding protein;PDBTitle: crystal structure of apo-ttha1159
86 d1y0ja1
not modelled
5.0
27
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Erythroid transcription factor GATA-1