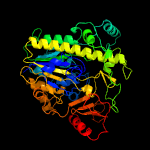
1 d1hdha_
100.0
33
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase
2 c4upiA_
100.0
21
PDB header: hydrolaseChain: A: PDB Molecule: sulfatase family protein;PDBTitle: dimeric sulfatase spas1 from silicibacter pomeroyi
3 c4uplC_
100.0
23
PDB header: hydrolaseChain: C: PDB Molecule: sulfatase family protein;PDBTitle: dimeric sulfatase spas2 from silicibacter pomeroyi
4 c3ed4A_
100.0
23
PDB header: transferaseChain: A: PDB Molecule: arylsulfatase;PDBTitle: crystal structure of putative arylsulfatase from escherichia coli
5 d1p49a_
100.0
23
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase
6 c4uphA_
100.0
22
PDB header: hydrolaseChain: A: PDB Molecule: sulfatase (sulfuric ester hydrolase) protein;PDBTitle: crystal structure of phosphonate monoester hydrolase of agrobacterium2 radiobacter
7 c4ug4H_
100.0
20
PDB header: hydrolaseChain: H: PDB Molecule: choline sulfatase;PDBTitle: crystal structure of a choline sulfatase from sinorhizobium2 melliloti
8 c4fdiA_
100.0
27
PDB header: hydrolaseChain: A: PDB Molecule: n-acetylgalactosamine-6-sulfatase;PDBTitle: the molecular basis of mucopolysaccharidosis iv a
9 c6b1vB_
100.0
27
PDB header: hydrolaseChain: B: PDB Molecule: iota-carrageenan sulfatase;PDBTitle: crystal structure of ps i-cgsb c78s in complex with i-neocarratetraose
10 c4upkC_
100.0
21
PDB header: hydrolaseChain: C: PDB Molecule: phosphonate monoester hydrolase;PDBTitle: phosphonate monoester hydrolase sppmh from silicibacter pomeroyi
11 d1fsua_
100.0
29
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase
12 c2qzuA_
100.0
21
PDB header: hydrolaseChain: A: PDB Molecule: putative sulfatase yidj;PDBTitle: crystal structure of the putative sulfatase yidj from bacteroides2 fragilis. northeast structural genomics consortium target bfr123
13 d1auka_
100.0
27
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase
14 c5fqlA_
100.0
19
PDB header: hydrolaseChain: A: PDB Molecule: iduronate-2-sulfatase;PDBTitle: insights into hunter syndrome from the structure of iduronate-2-2 sulfatase
15 c6j66B_
100.0
23
PDB header: hydrolaseChain: B: PDB Molecule: chondroitin sulfate/dermatan sulfate 4-o-endosulfatasePDBTitle: chondroitin sulfate/dermatan sulfate endolytic 4-o-sulfatase
16 c4mivB_
100.0
24
PDB header: hydrolaseChain: B: PDB Molecule: n-sulphoglucosamine sulphohydrolase;PDBTitle: crystal structure of sulfamidase, crystal form l
17 c2vqrA_
100.0
21
PDB header: hydrolaseChain: A: PDB Molecule: putative sulfatase;PDBTitle: crystal structure of a phosphonate monoester hydrolase from rhizobium2 leguminosarum: a new member of the alkaline phosphatase superfamily
18 c3b5qB_
100.0
21
PDB header: hydrolaseChain: B: PDB Molecule: putative sulfatase yidj;PDBTitle: crystal structure of a putative sulfatase (np_810509.1) from2 bacteroides thetaiotaomicron vpi-5482 at 2.40 a resolution
19 c6hhmA_
100.0
22
PDB header: hydrolaseChain: A: PDB Molecule: arylsulfatase;PDBTitle: crystal structure of the family s1_7 ulvan-specific sulfatase fa220702 from formosa agariphila
20 c5g2vA_
100.0
27
PDB header: hydrolaseChain: A: PDB Molecule: n-acetylglucosamine-6-sulfatase;PDBTitle: structure of bt4656 in complex with its substrate d-glucosamine-2-n,2 6-o-disulfate.
21 c6hr5A_
not modelled
100.0
24
PDB header: hydrolaseChain: A: PDB Molecule: alpha-l-rhamnosidase/sulfatase (gh78);PDBTitle: structure of the s1_25 family sulfatase module of the rhamnosidase2 fa22250 from formosa agariphila
22 c4uopB_
not modelled
100.0
16
PDB header: transferaseChain: B: PDB Molecule: lipoteichoic acid primase;PDBTitle: crystal structure of the lipoteichoic acid synthase ltap from listeria2 monocytogenes
23 c3lxqB_
not modelled
100.0
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein vp1736;PDBTitle: the crystal structure of a protein in the alkaline phosphatase2 superfamily from vibrio parahaemolyticus to 1.95a
24 c2w8dB_
not modelled
100.0
17
PDB header: transferaseChain: B: PDB Molecule: processed glycerol phosphate lipoteichoic acid synthase 2;PDBTitle: distinct and essential morphogenic functions for wall- and2 lipo-teichoic acids in bacillus subtilis
25 c2w5tA_
not modelled
100.0
16
PDB header: transferaseChain: A: PDB Molecule: processed glycerol phosphate lipoteichoic acidPDBTitle: structure-based mechanism of lipoteichoic acid synthesis by2 staphylococcus aureus ltas.
26 c4uorK_
not modelled
100.0
15
PDB header: transferaseChain: K: PDB Molecule: lipoteichoic acid synthase;PDBTitle: structure of lipoteichoic acid synthase ltas from listeria2 monocytogenes in complex with glycerol phosphate
27 c5i5fA_
not modelled
100.0
21
PDB header: membrane proteinChain: A: PDB Molecule: inner membrane protein yejm;PDBTitle: salmonella global domain 191
28 c2zktB_
not modelled
100.0
16
PDB header: isomeraseChain: B: PDB Molecule: 2,3-bisphosphoglycerate-independent phosphoglyceratePDBTitle: structure of ph0037 protein from pyrococcus horikoshii
29 c5k4pA_
not modelled
100.0
16
PDB header: transferaseChain: A: PDB Molecule: probable phosphatidylethanolamine transferase mcr-1;PDBTitle: catalytic domain of mcr-1 phosphoethanolamine transferase
30 c6bneA_
not modelled
100.0
15
PDB header: transferaseChain: A: PDB Molecule: phosphoethanolamine transferase;PDBTitle: crystal structure of the intrinsic colistin resistance enzyme icr(mc)2 from moraxella catarrhalis, catalytic domain, phosphate-bound complex
31 c3m8yC_
not modelled
100.0
22
PDB header: isomeraseChain: C: PDB Molecule: phosphopentomutase;PDBTitle: phosphopentomutase from bacillus cereus after glucose-1,6-bisphosphate2 activation
32 c4kayA_
not modelled
100.0
18
PDB header: transferaseChain: A: PDB Molecule: yhbx/yhjw/yijp/yjdb family protein;PDBTitle: structure of the soluble domain of lipooligosaccharide2 phosphoethanolamine transferase a from neisseria meningitidis -3 complex with zn
33 c5tj3A_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: alkaline phosphatase pafa;PDBTitle: crystal structure of wild type alkaline phosphatase pafa to 1.7a2 resolution
34 c6a82A_
not modelled
100.0
14
PDB header: transferaseChain: A: PDB Molecule: phosphoethanolamine transferase eptc;PDBTitle: crystal structure of the c-terminal periplasmic domain of eceptc from2 escherichia coli
35 c3q3qA_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: alkaline phosphatase;PDBTitle: crystal structure of spap: an novel alkaline phosphatase from2 bacterium sphingomonas sp. strain bsar-1
36 c4tn0C_
not modelled
100.0
15
PDB header: transferaseChain: C: PDB Molecule: upf0141 protein yjdb;PDBTitle: crystal structure of the c-terminal periplasmic domain of2 phosphoethanolamine transferase eptc from campylobacter jejuni
37 c2i09A_
not modelled
100.0
18
PDB header: isomeraseChain: A: PDB Molecule: phosphopentomutase;PDBTitle: crystal structure of putative phosphopentomutase from streptococcus2 mutans
38 c4lqyA_
not modelled
100.0
14
PDB header: hydrolaseChain: A: PDB Molecule: bis(5'-adenosyl)-triphosphatase enpp4;PDBTitle: crystal structure of human enpp4 with amp
39 c5udyA_
not modelled
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: human alkaline sphingomyelinase (alk-smase, enpp7, npp7)
40 c2gsoB_
not modelled
100.0
17
PDB header: hydrolaseChain: B: PDB Molecule: phosphodiesterase-nucleotide pyrophosphatase;PDBTitle: structure of xac nucleotide pyrophosphatase/phosphodiesterase in2 complex with vanadate
41 d1o98a2
not modelled
100.0
20
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, catalytic domain
42 c5vemA_
not modelled
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: human ectonucleotide pyrophosphatase / phosphodiesterase 5 (enpp5,2 npp5)
43 c5egeD_
not modelled
100.0
12
PDB header: hydrolaseChain: D: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: structure of enpp6, a choline-specific glycerophosphodiester-2 phosphodiesterase
44 c5u9zB_
not modelled
100.0
15
PDB header: transferaseChain: B: PDB Molecule: phosphoglycerol transferase;PDBTitle: phosphoglycerol transferase gach from streptococcus pyogenes
45 c3szzA_
not modelled
100.0
17
PDB header: hydrolaseChain: A: PDB Molecule: phosphonoacetate hydrolase;PDBTitle: crystal structure of phosphonoacetate hydrolase from sinorhizobium2 meliloti 1021 in complex with acetate
46 d2i09a1
not modelled
100.0
24
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: DeoB catalytic domain-like
47 d1ei6a_
not modelled
100.0
16
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Phosphonoacetate hydrolase
48 c5fgnA_
not modelled
100.0
20
PDB header: transferase,hydrolaseChain: A: PDB Molecule: lipooligosaccharide phosphoethanolamine transferase a;PDBTitle: integral membrane protein lipooligosaccharide phosphoethanolamine2 transferase a (epta) from neisseria meningitidis
49 c6c02B_
not modelled
100.0
13
PDB header: hydrolaseChain: B: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: human ectonucleotide pyrophosphatase / phosphodiesterase 3 (enpp3,2 npp3, cd203c), inactive (t205a), n594s, with alpha,beta-methylene-atp3 (ampcpp)
50 c5gz5A_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: snake venom phosphodiesterase (pde);PDBTitle: crystal structure of snake venom phosphodiesterase (pde) from taiwan2 cobra (naja atra atra) in complex with amp
51 c5gz4A_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: snake venom phosphodiesterase (pde);PDBTitle: crystal structure of snake venom phosphodiesterase (pde) from taiwan2 cobra (naja atra atra)
52 c4b56A_
not modelled
100.0
18
PDB header: hydrolaseChain: A: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: structure of ectonucleotide pyrophosphatase-phosphodiesterase-12 (npp1)
53 c2xrgA_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: crystal structure of autotaxin (enpp2) in complex with the2 ha155 boronic acid inhibitor
54 c2xr9A_
not modelled
100.0
15
PDB header: hydrolaseChain: A: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: crystal structure of autotaxin (enpp2)
55 c1o98A_
not modelled
99.8
25
PDB header: isomeraseChain: A: PDB Molecule: 2,3-bisphosphoglycerate-independentPDBTitle: 1.4a crystal structure of phosphoglycerate mutase from2 bacillus stearothermophilus complexed with3 2-phosphoglycerate
56 c5kgmA_
not modelled
99.8
25
PDB header: isomeraseChain: A: PDB Molecule: 2,3-bisphosphoglycerate-independent phosphoglyceratePDBTitle: 2.95a resolution structure of apo independent phosphoglycerate mutase2 from c. elegans (monoclinic form)
57 c3igzB_
not modelled
99.7
22
PDB header: isomeraseChain: B: PDB Molecule: cofactor-independent phosphoglycerate mutase;PDBTitle: crystal structures of leishmania mexicana phosphoglycerate2 mutase at low cobalt concentration
58 c2d1gB_
not modelled
99.7
18
PDB header: hydrolaseChain: B: PDB Molecule: acid phosphatase;PDBTitle: structure of francisella tularensis acid phosphatase a (acpa) bound to2 orthovanadate
59 c5vpuA_
not modelled
99.7
18
PDB header: isomeraseChain: A: PDB Molecule: 2,3-bisphosphoglycerate-independent phosphoglyceratePDBTitle: crystal structure of 2,3-bisphosphoglycerate-independent2 phosphoglycerate mutase bound to 3-phosphoglycerate, from3 acinetobacter baumannii
60 c4my4A_
not modelled
99.7
19
PDB header: isomeraseChain: A: PDB Molecule: 2,3-bisphosphoglycerate-independent phosphoglyceratePDBTitle: crystal structure of phosphoglycerate mutase from staphylococcus2 aureus.
61 c2iucB_
not modelled
99.4
16
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: structure of alkaline phosphatase from the antarctic bacterium tab5
62 d1k7ha_
not modelled
99.3
16
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Alkaline phosphatase
63 d1y6va1
not modelled
99.3
22
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Alkaline phosphatase
64 c1ew2A_
not modelled
99.3
18
PDB header: hydrolaseChain: A: PDB Molecule: phosphatase;PDBTitle: crystal structure of a human phosphatase
65 d1zeda1
not modelled
99.2
18
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Alkaline phosphatase
66 c2w0yB_
not modelled
99.0
17
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: h.salinarum alkaline phosphatase
67 c3a52A_
not modelled
99.0
18
PDB header: hydrolaseChain: A: PDB Molecule: cold-active alkaline phosphatase;PDBTitle: crystal structure of cold-active alkailne phosphatase from2 psychrophile shewanella sp.
68 c2x98A_
not modelled
99.0
18
PDB header: hydrolaseChain: A: PDB Molecule: alkaline phosphatase;PDBTitle: h.salinarum alkaline phosphatase
69 c3e2dB_
not modelled
98.9
19
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: the 1.4 a crystal structure of the large and cold-active vibrio sp.2 alkaline phosphatase
70 c3wbhB_
not modelled
98.9
18
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: structural characteristics of alkaline phosphatase from a moderately2 halophilic bacteria halomonas sp.593
71 c2r1dF_
not modelled
98.2
20
PDB header: cell adhesion, splicingChain: F: PDB Molecule: neurexin-1-beta;PDBTitle: crystal structure of rat neurexin 1beta in the ca2+ containing form
72 c3asiA_
not modelled
98.1
22
PDB header: cell adhesionChain: A: PDB Molecule: neurexin-1-alpha;PDBTitle: alpha-neurexin-1 ectodomain fragment; lns5-egf3-lns6
73 c3poyA_
not modelled
98.1
19
PDB header: cell adhesionChain: A: PDB Molecule: neurexin-1-alpha;PDBTitle: crystal structure of the alpha-neurexin-1 ectodomain, lns 2-6
74 c1okqA_
not modelled
98.0
14
PDB header: metal binding proteinChain: A: PDB Molecule: laminin alpha 2 chain;PDBTitle: laminin alpha 2 chain lg4-5 domain pair, ca1 site mutant
75 c2jkbA_
not modelled
98.0
16
PDB header: lyaseChain: A: PDB Molecule: sialidase b;PDBTitle: crystal structure of streptococcus pneumoniae nanb in2 complex with 2,7-anhydro-neu5ac
76 c2jd4B_
not modelled
98.0
17
PDB header: metal binding proteinChain: B: PDB Molecule: laminin subunit alpha-1;PDBTitle: mouse laminin alpha1 chain, domains lg4-5
77 c3qcwB_
not modelled
98.0
19
PDB header: cell adhesionChain: B: PDB Molecule: neurexin-1-alpha;PDBTitle: structure of neurexin 1 alpha (domains lns1-lns6), no splice inserts
78 d2erfa1
not modelled
97.9
16
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Laminin G-like module
79 d1dyka1
not modelled
97.9
13
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Laminin G-like module
80 c3v65A_
not modelled
97.9
16
PDB header: protein bindingChain: A: PDB Molecule: agrin;PDBTitle: crystal structure of agrin and lrp4 complex
81 c4yw5A_
not modelled
97.8
13
PDB header: hydrolaseChain: A: PDB Molecule: neuraminidase c;PDBTitle: crystal structure of streptococcus pneumoniae nanc, complex with2 oseltamivir carboxylate
82 c1za4A_
not modelled
97.8
16
PDB header: cell adhesionChain: A: PDB Molecule: thrombospondin 1;PDBTitle: crystal structure of the thrombospondin-1 n-terminal domain2 in complex with arixtra
83 c3flpJ_
not modelled
97.8
13
PDB header: sugar binding proteinChain: J: PDB Molecule: sap-like pentraxin;PDBTitle: crystal structure of native heptameric sap-like pentraxin2 from limulus polyphemus
84 c2wjsA_
not modelled
97.8
15
PDB header: cell adhesionChain: A: PDB Molecule: laminin subunit alpha-2;PDBTitle: crystal structure of the lg1-3 region of the laminin alpha22 chain
85 c2v73B_
not modelled
97.8
11
PDB header: sugar-binding proteinChain: B: PDB Molecule: putative exo-alpha-sialidase;PDBTitle: the structure of the family 40 cbm from c. perfringens nanj2 in complex with a sialic acid containing molecule
86 d2r1da1
not modelled
97.7
20
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Laminin G-like module
87 c2h0bC_
not modelled
97.7
15
PDB header: cell adhesionChain: C: PDB Molecule: neurexin-1-alpha;PDBTitle: crystal structure of the second lns/lg domain from neurexin 1 alpha
88 c3vkfD_
not modelled
97.7
20
PDB header: cell adhesionChain: D: PDB Molecule: neurexin-1-beta;PDBTitle: crystal structure of neurexin 1beta/neuroligin 1 complex
89 c3vkfC_
not modelled
97.7
20
PDB header: cell adhesionChain: C: PDB Molecule: neurexin-1-beta;PDBTitle: crystal structure of neurexin 1beta/neuroligin 1 complex
90 c5fraD_
not modelled
97.6
10
PDB header: sugar binding proteinChain: D: PDB Molecule: sialidase;PDBTitle: cbm40_cpf0721-6'sl
91 c6er3B_
not modelled
97.6
17
PDB header: sugar binding proteinChain: B: PDB Molecule: bnr/asp-box repeat protein;PDBTitle: ruminococcus gnavus it-sialidase cbm40 bound to alpha2,3 sialyllactose
92 c2c5dA_
not modelled
97.6
17
PDB header: signaling protein/receptorChain: A: PDB Molecule: growth-arrest-specific protein 6 precursor;PDBTitle: structure of a minimal gas6-axl complex
93 c3mw3A_
not modelled
97.6
17
PDB header: cell adhesionChain: A: PDB Molecule: neurexin-2-beta;PDBTitle: crystal structure of beta-neurexin 2 with the splice insert 4
94 c2r16A_
not modelled
97.5
18
PDB header: cell adhesion, splicingChain: A: PDB Molecule: neurexin-1-alpha;PDBTitle: crystal structure of bovine neurexin 1 alpha lns/lg domain 4 (with no2 splice insert)
95 c5mc9A_
not modelled
97.5
15
PDB header: cell adhesionChain: A: PDB Molecule: laminin subunit alpha-1;PDBTitle: crystal structure of the heterotrimeric integrin-binding region of2 laminin-111
96 d1saca_
not modelled
97.3
18
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Pentraxin (pentaxin)
97 d2slia1
not modelled
97.3
14
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Leech intramolecular trans-sialidase, N-terminal domain
98 d1q56a_
not modelled
97.2
15
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Laminin G-like module
99 c3pveA_
not modelled
97.2
18
PDB header: transcriptionChain: A: PDB Molecule: agrin, agrin protein;PDBTitle: crystal structure of the g2 domain of agrin from mus musculus
100 c1fv3B_
not modelled
97.2
11
PDB header: toxinChain: B: PDB Molecule: tetanus toxin heavy chain;PDBTitle: the hc fragment of tetanus toxin complexed with an analogue of its2 ganglioside receptor gt1b
101 c5oltA_
not modelled
97.2
15
PDB header: transferaseChain: A: PDB Molecule: cellulose biosynthesis protein bcsg;PDBTitle: crystal structure of the extramembrane domain of the cellulose2 biosynthetic protein bcsg from salmonella typhimurium
102 c1yynA_
not modelled
97.2
10
PDB header: hydrolaseChain: A: PDB Molecule: tetanus toxin;PDBTitle: a common binding site for disialyllactose and a tri-peptide2 in the c-fragment of tetanus neurotoxin
103 c2sliA_
not modelled
97.1
15
PDB header: hydrolaseChain: A: PDB Molecule: intramolecular trans-sialidase;PDBTitle: leech intramolecular trans-sialidase complexed with 2,7-2 anhydro-neu5ac, the reaction product
104 c1qu0A_
not modelled
97.1
20
PDB header: metal binding proteinChain: A: PDB Molecule: laminin alpha2 chain;PDBTitle: crystal structure of the fifth laminin g-like module of the2 mouse laminin alpha2 chain
105 c1qu0D_
not modelled
97.1
20
PDB header: metal binding proteinChain: D: PDB Molecule: laminin alpha2 chain;PDBTitle: crystal structure of the fifth laminin g-like module of the2 mouse laminin alpha2 chain
106 d1h30a1
not modelled
97.1
23
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Laminin G-like module
107 c3sh5A_
not modelled
97.1
17
PDB header: metal binding proteinChain: A: PDB Molecule: lg3 peptide;PDBTitle: calcium-bound laminin g like domain 3 from human perlecan
108 c4dqaA_
not modelled
97.1
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative carbohydrate binding protein2 (bacova_03559) from bacteroides ovatus atcc 8483 at 1.50 a resolution
109 c2vxrA_
not modelled
97.1
8
PDB header: toxinChain: A: PDB Molecule: botulinum neurotoxin type g;PDBTitle: crystal structure of the botulinum neurotoxin serotype g2 binding domain
110 d1b09a_
not modelled
97.0
17
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Pentraxin (pentaxin)
111 d1pz7a_
not modelled
97.0
16
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Laminin G-like module
112 c3mppG_
not modelled
97.0
9
PDB header: toxinChain: G: PDB Molecule: botulinum neurotoxin type g;PDBTitle: botulinum neurotoxin type g receptor binding domain
113 c4pboA_
not modelled
97.0
16
PDB header: immune systemChain: A: PDB Molecule: c-reactive protein;PDBTitle: crystal structure of zebrafish short-chain pentraxin protein without2 calcium ions
114 d1h30a2
not modelled
96.9
18
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Laminin G-like module
115 c5ho0A_
not modelled
96.9
16
PDB header: hydrolaseChain: A: PDB Molecule: extracellular arabinanase;PDBTitle: crystal structure of abna (closed conformation), a gh43 extracellular2 arabinanase from geobacillus stearothermophilus
116 c1qu0C_
not modelled
96.8
20
PDB header: metal binding proteinChain: C: PDB Molecule: laminin alpha2 chain;PDBTitle: crystal structure of the fifth laminin g-like module of the2 mouse laminin alpha2 chain
117 c6hoxA_
not modelled
96.7
10
PDB header: toxinChain: A: PDB Molecule: binding domain (hc) of paraclostridial mosquitocidalPDBTitle: crystal structure of the binding domain of paraclostridial2 mosquitocidal protein 1
118 c4qvsA_
not modelled
96.7
13
PDB header: unknown functionChain: A: PDB Molecule: s-layer domain-containing protein;PDBTitle: 2.1 angstrom resolution crystal structure of s-layer domain-containing2 protein (residues 221-444) from clostridium thermocellum atcc 27405
119 c3fuqA_
not modelled
96.6
12
PDB header: toxinChain: A: PDB Molecule: bont/f (neurotoxin type f);PDBTitle: glycosylated sv2 and gangliosides as dual receptors for2 botulinum neurotoxin serotype f
120 c2vu9A_
not modelled
96.6
11
PDB header: hydrolaseChain: A: PDB Molecule: botulinum neurotoxin a heavy chain;PDBTitle: crystal structure of botulinum neurotoxin serotype a2 binding domain in complex with gt1b