1 c5o60F_
100.0
84
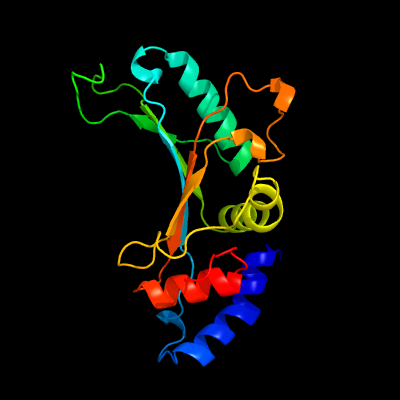
PDB header: ribosomeChain: F: PDB Molecule: 50s ribosomal protein l5;PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
2 d1iq4a_
100.0
57
Fold: RL5-likeSuperfamily: RL5-likeFamily: Ribosomal protein L5
3 d1mjia_
100.0
58
Fold: RL5-likeSuperfamily: RL5-likeFamily: Ribosomal protein L5
4 d2zjrd1
100.0
54
Fold: RL5-likeSuperfamily: RL5-likeFamily: Ribosomal protein L5
5 c3bboH_
100.0
49
PDB header: ribosomeChain: H: PDB Molecule: ribosomal protein l5;PDBTitle: homology model for the spinach chloroplast 50s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome
6 d2gycd1
100.0
57
Fold: RL5-likeSuperfamily: RL5-likeFamily: Ribosomal protein L5
7 c1vw4E_
100.0
38
PDB header: ribosomeChain: E: PDB Molecule: 54s ribosomal protein l7, mitochondrial;PDBTitle: structure of the yeast mitochondrial large ribosomal subunit
8 c3j21E_
100.0
30
PDB header: ribosomeChain: E: PDB Molecule: 50s ribosomal protein l5p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
9 c4a1cD_
100.0
31
PDB header: ribosomeChain: D: PDB Molecule: 60s ribosomal protein l11;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
10 c3zf7L_
100.0
27
PDB header: ribosomeChain: L: PDB Molecule: 60s ribosomal protein l11, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
11 c1s1iJ_
100.0
31
PDB header: ribosomeChain: J: PDB Molecule: 60s ribosomal protein l11;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1i, contains 60s subunit. the 40s4 ribosomal subunit is in file 1s1h.
12 d1vqod1
100.0
42
Fold: RL5-likeSuperfamily: RL5-likeFamily: Ribosomal protein L5
13 c4xeoB_
63.8
22
PDB header: ligaseChain: B: PDB Molecule: alanine--trna ligase, cytoplasmic;PDBTitle: crystal structure of human alars catalytic domain with r329h mutation
14 c5knnG_
57.0
22
PDB header: ligaseChain: G: PDB Molecule: alanine--trna ligase, cytoplasmic;PDBTitle: evolutionary gain of alanine mischarging to non-cognate trnas with a2 g4:u69 base pair
15 c3hxxA_
26.6
22
PDB header: ligaseChain: A: PDB Molecule: alanyl-trna synthetase;PDBTitle: crystal structure of catalytic fragment of e. coli alars in complex2 with amppcp
16 c2rf4B_
23.2
45
PDB header: transferaseChain: B: PDB Molecule: dna-directed rna polymerase i subunit rpa4;PDBTitle: crystal structure of the rna polymerase i subcomplex a14/43
17 c1yfsB_
21.6
22
PDB header: ligaseChain: B: PDB Molecule: alanyl-trna synthetase;PDBTitle: the crystal structure of alanyl-trna synthetase in complex2 with l-alanine
18 c5o8oA_
18.2
23
PDB header: protein transportChain: A: PDB Molecule: mitochondrial import receptor subunit tom40;PDBTitle: n. crassa tom40 model based on cryo-em structure of the tom core2 complex at 6.8 a
19 c2jvfA_
13.9
24
PDB header: de novo proteinChain: A: PDB Molecule: de novo protein m7;PDBTitle: solution structure of m7, a computationally-designed2 artificial protein
20 d3d37a1
13.9
27
Fold: Phage tail proteinsSuperfamily: Phage tail proteinsFamily: Baseplate protein-like
21 c1qysA_
not modelled
13.6
19
PDB header: de novo proteinChain: A: PDB Molecule: top7;PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold
22 c4whjA_
not modelled
12.7
20
PDB header: antiviral protein, hydrolaseChain: A: PDB Molecule: interferon-induced gtp-binding protein mx2;PDBTitle: myxovirus resistance protein 2 (mxb)
23 d1riqa2
not modelled
12.7
22
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain
24 c4arjB_
not modelled
12.5
40
PDB header: hydrolaseChain: B: PDB Molecule: pesticin, lysozyme;PDBTitle: crystal structure of a pesticin (translocation and receptor binding2 domain) from y. pestis and t4-lysozyme chimera
25 c1xtzA_
not modelled
12.5
8
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase;PDBTitle: crystal structure of the s. cerevisiae d-ribose-5-phosphate isomerase:2 comparison with the archeal and bacterial enzymes
26 c4ky3A_
not modelled
10.6
19
PDB header: de novo proteinChain: A: PDB Molecule: designed protein or327;PDBTitle: three-dimensional structure of the orthorhombic crystal of2 computationally designed insertion domain , northeast structural3 genomics consortium (nesg) target or327
27 c4c8qA_
not modelled
9.7
27
PDB header: transcriptionChain: A: PDB Molecule: sm-like protein lsm1;PDBTitle: crystal structure of the yeast lsm1-7-pat1 complex
28 c3j3v5_
not modelled
9.4
23
PDB header: ribosomeChain: 5: PDB Molecule: 50s ribosomal protein l1;PDBTitle: atomic model of the immature 50s subunit from bacillus subtilis (state2 i-a)
29 d1a8ya3
not modelled
9.0
16
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Calsequestrin
30 d2b8ea1
not modelled
8.5
17
Fold: HAD-likeSuperfamily: HAD-likeFamily: Meta-cation ATPase, catalytic domain P
31 d1wmxa_
not modelled
7.8
16
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 30 carbohydrate binding module, CBM30 (PKD repeat)
32 d1j6xa_
not modelled
7.7
9
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: Autoinducer-2 production protein LuxS
33 c2ebbA_
not modelled
7.3
17
PDB header: lyaseChain: A: PDB Molecule: pterin-4-alpha-carbinolamine dehydratase;PDBTitle: crystal structure of pterin-4-alpha-carbinolamine2 dehydratase (pterin carbinolamine dehydratase) from3 geobacillus kaustophilus hta426
34 c5owgA_
not modelled
7.2
21
PDB header: oxidoreductaseChain: A: PDB Molecule: pcyx_ebk42635;PDBTitle: structure of pcyx_ebk42635
35 d1a62a2
not modelled
6.6
25
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
36 c2xznH_
not modelled
6.2
26
PDB header: ribosomeChain: H: PDB Molecule: ribosomal protein s8 containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
37 d1nt2b_
not modelled
5.9
21
Fold: Nop domainSuperfamily: Nop domainFamily: Nop domain
38 d1i6ua_
not modelled
5.8
20
Fold: Ribosomal protein S8Superfamily: Ribosomal protein S8Family: Ribosomal protein S8
39 d3eipa_
not modelled
5.8
50
Fold: FKBP-likeSuperfamily: Colicin E3 immunity proteinFamily: Colicin E3 immunity protein
40 c2x9oA_
not modelled
5.8
13
PDB header: oxidoreductaseChain: A: PDB Molecule: 15,16-dihydrobiliverdin-ferredoxin oxidoreductase;PDBTitle: structure of 15, 16- dihydrobiliverdin:ferredoxin2 oxidoreductase (peba)
41 c2x6vB_
not modelled
5.7
18
PDB header: transcription/dnaChain: B: PDB Molecule: t-box transcription factor tbx5;PDBTitle: crystal structure of human tbx5 in the dna-bound and dna-2 free form
42 c5el3D_
not modelled
5.4
11
PDB header: rna binding proteinChain: D: PDB Molecule: kh domain-containing, rna-binding, signal transduction-PDBTitle: structure of the kh domain of t-star
43 d3cjsb1
not modelled
5.4
27
Fold: Ribosomal L11/L12e N-terminal domainSuperfamily: Ribosomal L11/L12e N-terminal domainFamily: Ribosomal L11/L12e N-terminal domain
44 c6em5J_
not modelled
5.4
35
PDB header: ribosomeChain: J: PDB Molecule: rrna-processing protein ebp2;PDBTitle: state d architectural model (nsa1-tap flag-ytm1) - visualizing the2 assembly pathway of nucleolar pre-60s ribosomes
45 c2g18K_
not modelled
5.3
15
PDB header: oxidoreductaseChain: K: PDB Molecule: phycocyanobilin:ferredoxin oxidoreductase;PDBTitle: crystal structure of nostoc sp. 7120 phycocyanobilin:ferredoxin2 oxidoreductase (pcya) apoprotein
46 c4walA_
not modelled
5.3
22
PDB header: protein binding/rnaChain: A: PDB Molecule: branchpoint-bridging protein;PDBTitle: crystal structure of selenomethionine msl5 protein in complex with rna2 at 2.2 a
47 d1k1ga_
not modelled
5.2
22
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)
48 d1mmsa2
not modelled
5.2
27
Fold: Ribosomal L11/L12e N-terminal domainSuperfamily: Ribosomal L11/L12e N-terminal domainFamily: Ribosomal L11/L12e N-terminal domain