|
|
 |
| Summary |
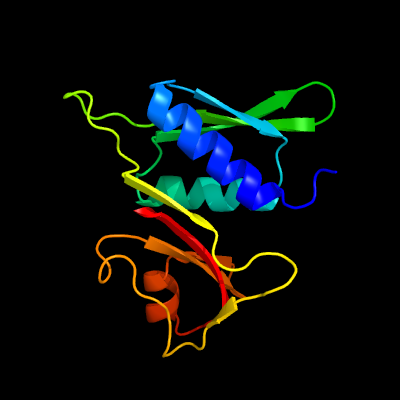
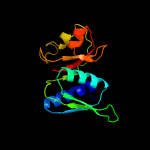
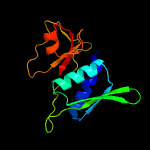
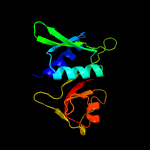
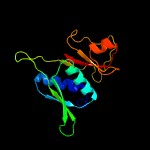
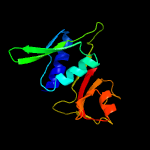
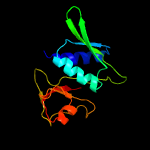
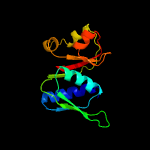
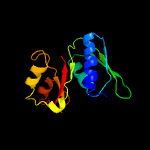
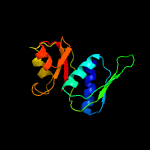
| Top model | |||||||||||||||
| |||||||||||||||
| Sequence analysis |
| Download FASTA version |
| Secondary structure and disorder prediction [Show] |
 |
 |
| 1 | . | . | . | . | . | . | . | . | 10 | . | . | . | . | . | . | . | . | . | 20 | . | . | . | . | . | . | . | . | . | 30 | . | . | . | . | . | . | . | . | . | 40 | . | . | . | . | . | . | . | . | . | 50 | . | . | . | . | . | . | . | . | . | 60 | |||||||||||||||||||||
| Sequence | M | T | M | T | D | P | I | A | D | F | L | T | R | L | R | N | A | N | S | A | Y | H | D | E | V | S | L | P | H | S | K | L | K | A | N | I | A | Q | I | L | K | N | E | G | Y | I | S | D | F | R | T | E | D | A | R | V | G | K | S | L | ||||||||||||||||||||
| Secondary structure |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  | ||||||||||||||||||||||||||||||||||||||
| SS confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Disorder | ? | ? | ? | ? | ? | ? | ? | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Disorder confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| . | . | . | . | . | . | . | . | . | 70 | . | . | . | . | . | . | . | . | . | 80 | . | . | . | . | . | . | . | . | . | 90 | . | . | . | . | . | . | . | . | . | 100 | . | . | . | . | . | . | . | . | . | 110 | . | . | . | . | . | . | . | . | . | 120 | |||||||||||||||||||||
| Sequence | V | I | Q | L | K | Y | G | P | S | R | E | R | S | I | A | G | L | R | R | V | S | K | P | G | L | R | V | Y | A | K | S | T | N | L | P | R | V | L | G | G | L | G | V | A | I | I | S | T | S | S | G | L | L | T | D | R | Q | A | A | R | ||||||||||||||||||||
| Secondary structure |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  | ||||||||||||||||||||||||||||||||||||||||||||
| SS confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Disorder | ? | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Disorder confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| . | . | . | . | . | . | . | . | . | 130 | . | . | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sequence | Q | G | V | G | G | E | V | L | A | Y | V | W | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary structure |  |  |  |  |  |  | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SS confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Disorder | ? | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Disorder confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||
| Domain analysis [Show] |
 |
 |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
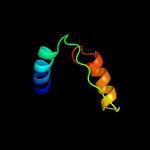
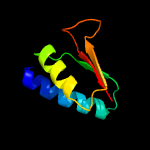
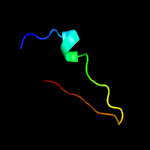
| Detailed template information [Hide] |
| Binding site prediction |
Due to computational demand, binding site predictions are not run for batch jobs
If you want to predict binding sites, please manually submit your model of choice to 3DLigandSite
Phyre is now FREE for commercial users!
All images and data generated by Phyre2 are free to use in any publication with acknowledgement
| Please cite: The Phyre2 web portal for protein modeling, prediction and analysis. | ||||||||||||||
| Kelley LA et al.. Nature Protocols 10, 845-858 (2015)[pdf] [Citation link] | ||||||||||||||
| If you use the binding site predictions from 3DLigandSite, please also cite: | ||||||||||||||
| 3DLigandSite: predicting ligand-binding sites using similar structures. | ||||||||||||||
| Wass MN, Kelley LA and Sternberg MJ Nucleic Acids Research 38, W469-73 (2010) [PubMed] | ||||||||||||||
|
|
| ||||||||||||