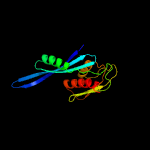
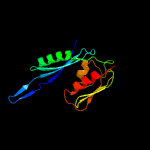
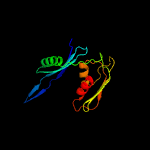
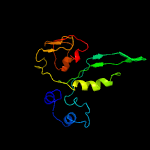
1 c5o5jE_
100.0
96
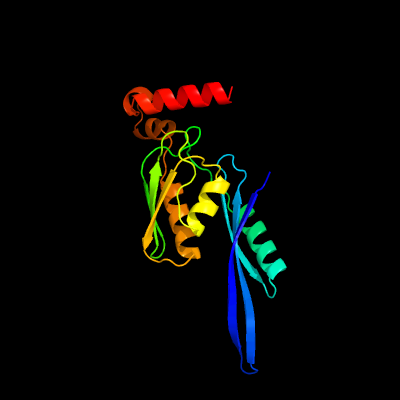
PDB header: ribosomeChain: E: PDB Molecule: 30s ribosomal protein s5;PDBTitle: structure of the 30s small ribosomal subunit from mycobacterium2 smegmatis
2 c3j6vE_
100.0
21
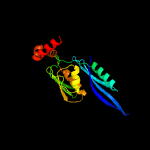
PDB header: ribosomeChain: E: PDB Molecule: 28s ribosomal protein s5, mitochondrial;PDBTitle: cryo-em structure of the small subunit of the mammalian mitochondrial2 ribosome
3 c5mmje_
100.0
42
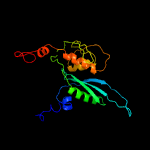
PDB header: ribosomeChain: E: PDB Molecule: PDBTitle: structure of the small subunit of the chloroplast ribosome
4 c3bbnE_
100.0
48
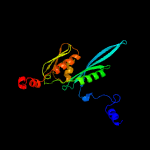
PDB header: ribosomeChain: E: PDB Molecule: ribosomal protein s5;PDBTitle: homology model for the spinach chloroplast 30s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome.
5 c1p6gE_
100.0
60
PDB header: ribosomeChain: E: PDB Molecule: 30s ribosomal protein s5;PDBTitle: real space refined coordinates of the 30s subunit fitted into the low2 resolution cryo-em map of the ef-g.gtp state of e. coli 70s ribosome
6 c2ow8f_
100.0
49
PDB header: ribosomeChain: F: PDB Molecule: PDBTitle: crystal structure of a 70s ribosome-trna complex reveals functional2 interactions and rearrangements. this file, 2ow8, contains the 30s3 ribosome subunit, two trna, and mrna molecules. 50s ribosome subunit4 is in the file 1vsa.
7 c2xzmE_
100.0
31
PDB header: ribosomeChain: E: PDB Molecule: ribosomal protein s5 containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
8 c1eg0B_
100.0
60
PDB header: ribosomePDB COMPND:
9 c3zeyP_
100.0
32
PDB header: ribosomeChain: P: PDB Molecule: 40s ribosomal protein s2, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
10 c4kzzC_
100.0
31
PDB header: ribosomeChain: C: PDB Molecule: 40s ribosomal protein s2;PDBTitle: rabbit 40s ribosomal subunit in complex with mrna, initiator trna and2 eif1a
11 c5xyiC_
100.0
31
PDB header: ribosomeChain: C: PDB Molecule: uncharacterized protein;PDBTitle: small subunit of trichomonas vaginalis ribosome
12 c3j20F_
100.0
29
PDB header: ribosomeChain: F: PDB Molecule: 30s ribosomal protein s5p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (30s3 ribosomal subunit)
13 c2zkqe_
100.0
32
PDB header: ribosomal protein/rnaChain: E: PDB Molecule: rna expansion segment es6 part ii;PDBTitle: structure of a mammalian ribosomal 40s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
14 c1s1hE_
100.0
34
PDB header: ribosomeChain: E: PDB Molecule: 40s ribosomal protein s2;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1h, contains 40s subunit. the 60s4 ribosomal subunit is in file 1s1i.
15 c3izbE_
100.0
32
PDB header: ribosomeChain: E: PDB Molecule: 40s ribosomal protein rps2 (s5p);PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
16 c3iz6E_
100.0
33
PDB header: ribosomeChain: E: PDB Molecule: 40s ribosomal protein s2 (s5p);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
17 d2qale1
100.0
58
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Translational machinery components
18 d2uube1
100.0
52
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Translational machinery components
19 d1pkpa1
99.9
60
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Translational machinery components
20 d2uube2
99.9
46
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Ribosomal S5 protein, N-terminal domain
21 d2qale2
not modelled
99.9
62
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Ribosomal S5 protein, N-terminal domain
22 d1pkpa2
not modelled
99.9
62
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Ribosomal S5 protein, N-terminal domain
23 c1vw45_
not modelled
68.8
14
PDB header: ribosomeChain: 5: PDB Molecule: 54s ribosomal protein l3, mitochondrial;PDBTitle: structure of the yeast mitochondrial large ribosomal subunit
24 d2b7ta1
not modelled
55.6
22
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)
25 d1p42a1
not modelled
55.5
27
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase LpxC
26 d2nuga2
not modelled
52.0
19
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)
27 c4oogC_
not modelled
51.5
21
PDB header: hydrolase/rnaChain: C: PDB Molecule: ribonuclease 3;PDBTitle: crystal structure of yeast rnase iii (rnt1p) complexed with the2 product of dsrna processing
28 c2go4A_
not modelled
51.1
27
PDB header: hydrolaseChain: A: PDB Molecule: udp-3-o-[3-hydroxymyristoyl] n-acetylglucosaminePDBTitle: crystal structure of aquifex aeolicus lpxc complexed with tu-514
29 c3nzkB_
not modelled
48.6
47
PDB header: hydrolaseChain: B: PDB Molecule: udp-3-o-[3-hydroxymyristoyl] n-acetylglucosaminePDBTitle: structure of lpxc from yersinia enterocolitica complexed with chir0902 inhibitor
30 c2vesA_
not modelled
47.2
33
PDB header: hydrolaseChain: A: PDB Molecule: udp-3-o-acyl-n-acetylglucosamine deacetylase;PDBTitle: crystal structure of lpxc from pseudomonas aeruginosa complexed with2 the potent bb-78485 inhibitor
31 d1bf4a_
not modelled
46.8
31
Fold: SH3-like barrelSuperfamily: Chromo domain-likeFamily: "Histone-like" proteins from archaea
32 c2mdrA_
not modelled
43.3
10
PDB header: hydrolaseChain: A: PDB Molecule: double-stranded rna-specific adenosine deaminase;PDBTitle: solution structure of the third double-stranded rna-binding domain2 (dsrbd3) of human adenosine-deaminase adar1
33 c2zqeA_
not modelled
43.0
37
PDB header: dna binding proteinChain: A: PDB Molecule: muts2 protein;PDBTitle: crystal structure of the smr domain of thermus thermophilus muts2
34 c6htuA_
not modelled
42.6
20
PDB header: rna binding proteinChain: A: PDB Molecule: double-stranded rna-binding protein staufen homolog 1;PDBTitle: structure of hstau1 dsrbd3-4 in complex with arf1 rna
35 c3adjA_
not modelled
41.7
21
PDB header: gene regulationChain: A: PDB Molecule: f21m12.9 protein;PDBTitle: structure of arabidopsis hyl1 and its molecular implications for mirna2 processing
36 c2ljhA_
not modelled
41.1
19
PDB header: hydrolaseChain: A: PDB Molecule: double-stranded rna-specific editase adar;PDBTitle: nmr structure of double-stranded rna-specific editase adar
37 d2b7va1
not modelled
39.1
23
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)
38 d2vapa2
not modelled
38.9
20
Fold: Bacillus chorismate mutase-likeSuperfamily: Tubulin C-terminal domain-likeFamily: Tubulin, C-terminal domain
39 d2pifa1
not modelled
38.7
19
Fold: PSTPO5379-likeSuperfamily: PSTPO5379-likeFamily: PSTPO5379-like
40 d1stua_
not modelled
38.2
20
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)
41 d1sr9a3
not modelled
37.5
18
Fold: 2-isopropylmalate synthase LeuA, allosteric (dimerisation) domainSuperfamily: 2-isopropylmalate synthase LeuA, allosteric (dimerisation) domainFamily: 2-isopropylmalate synthase LeuA, allosteric (dimerisation) domain
42 d1w5fa2
not modelled
36.5
28
Fold: Bacillus chorismate mutase-likeSuperfamily: Tubulin C-terminal domain-likeFamily: Tubulin, C-terminal domain
43 d1o0wa2
not modelled
36.5
20
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)
44 c5npgA_
not modelled
35.6
10
PDB header: rna binding proteinChain: A: PDB Molecule: loquacious, isoform f;PDBTitle: solution structure of drosophila melanogaster loquacious dsrbd1
45 d1ofua2
not modelled
35.6
19
Fold: Bacillus chorismate mutase-likeSuperfamily: Tubulin C-terminal domain-likeFamily: Tubulin, C-terminal domain
46 d1rq2a2
not modelled
34.0
20
Fold: Bacillus chorismate mutase-likeSuperfamily: Tubulin C-terminal domain-likeFamily: Tubulin, C-terminal domain
47 d1x47a1
not modelled
33.6
22
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)
48 c4gisB_
not modelled
33.2
18
PDB header: lyaseChain: B: PDB Molecule: enolase;PDBTitle: crystal structure of an enolase family member from vibrio harveyi2 (efi-target 501692) with homology to mannonate dehydratase, with mg,3 glycerol and dicarboxylates bound (mixed loops, space group i4122)
49 d1qu6a2
not modelled
32.3
14
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)
50 c2ltrA_
not modelled
32.1
25
PDB header: rna binding proteinChain: A: PDB Molecule: protein rde-4;PDBTitle: solution structure of rde-4(32-136)
51 c3adiC_
not modelled
31.1
15
PDB header: gene regulation/rnaChain: C: PDB Molecule: f21m12.9 protein;PDBTitle: structure of arabidopsis hyl1 and its molecular implications for mirna2 processing
52 d1tzza2
not modelled
29.2
32
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like
53 d1uila_
not modelled
29.1
8
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)
54 c2l3jA_
not modelled
25.4
27
PDB header: hydrolase/rnaChain: A: PDB Molecule: double-stranded rna-specific editase 1;PDBTitle: the solution structure of the adar2 dsrbm-rna complex reveals a2 sequence-specific read out of the minor groove
55 d1t4oa_
not modelled
23.4
23
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)
56 c1t4oA_
not modelled
23.4
23
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease iii;PDBTitle: crystal structure of rnt1p dsrbd
57 d2dixa1
not modelled
22.3
22
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)
58 c2fkpC_
not modelled
22.3
21
PDB header: isomeraseChain: C: PDB Molecule: n-acylamino acid racemase;PDBTitle: the mutant g127c-t313c of deinococcus radiodurans n-2 acylamino acid racemase
59 d1jpdx2
not modelled
21.7
18
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like
60 c3c4tA_
not modelled
19.8
20
PDB header: hydrolaseChain: A: PDB Molecule: endoribonuclease dicer;PDBTitle: structure of rnaseiiib and dsrna binding domains of mouse dicer
61 c2n3hA_
not modelled
19.8
21
PDB header: rna binding proteinChain: A: PDB Molecule: double-stranded rna-binding protein 4;PDBTitle: solution structure of drb4 dsrbd2 (viz. drb4(81-151))
62 c5b16A_
not modelled
19.5
21
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease 3,drosha,ribonuclease 3,drosha,ribonuclease 3;PDBTitle: x-ray structure of drosha in complex with the c-terminal tail of2 dgcr8.
63 d1r0ma2
not modelled
19.3
19
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like
64 d2f1da2
not modelled
18.3
15
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Imidazole glycerol phosphate dehydratase
65 d1jdfa2
not modelled
17.5
22
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like
66 d1x48a1
not modelled
17.4
28
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)
67 d1bqga2
not modelled
17.1
27
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like
68 c5n8lA_
not modelled
16.5
20
PDB header: rna binding proteinChain: A: PDB Molecule: risc-loading complex subunit tarbp2;PDBTitle: structure of trbp dsrbd 1 and 2 in complex with a 19 bp sirna (complex2 b)
69 d1azpa_
not modelled
16.0
30
Fold: SH3-like barrelSuperfamily: Chromo domain-likeFamily: "Histone-like" proteins from archaea
70 d1kn0a_
not modelled
15.4
22
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: The homologous-pairing domain of Rad52 recombinase
71 d1znna1
not modelled
15.1
21
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: PdxS-like
72 c1znnF_
not modelled
15.1
21
PDB header: biosynthetic proteinChain: F: PDB Molecule: plp synthase;PDBTitle: structure of the synthase subunit of plp synthase
73 c2n3gA_
not modelled
15.1
12
PDB header: rna binding proteinChain: A: PDB Molecule: double-stranded rna-binding protein 4;PDBTitle: solution structure of drb4 dsrbd1 (viz. drb4(1-72))
74 c1yywB_
not modelled
14.9
23
PDB header: hydrolase/rnaChain: B: PDB Molecule: ribonuclease iii;PDBTitle: crystal structure of rnase iii from aquifex aeolicus2 complexed with double stranded rna at 2.8-angstrom3 resolution
75 d1l0oc_
not modelled
14.7
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain
76 c1l0oC_
not modelled
14.7
21
PDB header: protein bindingChain: C: PDB Molecule: sigma factor;PDBTitle: crystal structure of the bacillus stearothermophilus anti-2 sigma factor spoiiab with the sporulation sigma factor3 sigmaf
77 c3adlA_
not modelled
14.7
15
PDB header: gene regulation/rnaChain: A: PDB Molecule: risc-loading complex subunit tarbp2;PDBTitle: structure of trbp2 and its molecule implications for mirna processing
78 d2dmya1
not modelled
14.0
11
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)
79 d1u7na_
not modelled
13.7
13
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: PlsX-like
80 d1di2a_
not modelled
13.4
21
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)
81 c6ezmL_
not modelled
13.4
15
PDB header: lyaseChain: L: PDB Molecule: imidazoleglycerol-phosphate dehydratase;PDBTitle: imidazoleglycerol-phosphate dehydratase from saccharomyces cerevisiae
82 d1t4na_
not modelled
13.0
23
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)
83 c1h2iG_
not modelled
12.8
22
PDB header: dna binding proteinChain: G: PDB Molecule: dna repair protein rad52 homolog;PDBTitle: human rad52 protein, n-terminal domain
84 d1t4lb_
not modelled
12.5
25
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)
85 d1uhza_
not modelled
12.4
21
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)
86 c6ci7A_
not modelled
11.7
14
PDB header: biosynthetic proteinChain: A: PDB Molecule: ycao;PDBTitle: the structure of ycao from methanopyrus kandleri bound with amppcp and2 mg2+
87 c2n5lA_
not modelled
11.7
25
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease zc3h12a;PDBTitle: regnase-1 c-terminal domain
88 c1w5fA_
not modelled
11.4
28
PDB header: cell divisionChain: A: PDB Molecule: cell division protein ftsz;PDBTitle: ftsz, t7 mutated, domain swapped (t. maritima)
89 c2gezE_
not modelled
11.4
13
PDB header: hydrolaseChain: E: PDB Molecule: l-asparaginase alpha subunit;PDBTitle: crystal structure of potassium-independent plant asparaginase
90 d1x49a1
not modelled
11.2
13
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)
91 c3rv0B_
not modelled
11.2
14
PDB header: rna binding proteinChain: B: PDB Molecule: k. polysporus dcr1;PDBTitle: crystal structure of k. polysporus dcr1 without the c-terminal dsrbd
92 d1tifa_
not modelled
11.0
53
Fold: beta-Grasp (ubiquitin-like)Superfamily: Translation initiation factor IF3, N-terminal domainFamily: Translation initiation factor IF3, N-terminal domain
93 c3u4fC_
not modelled
10.9
13
PDB header: isomeraseChain: C: PDB Molecule: mandelate racemase/muconate lactonizing enzyme familyPDBTitle: crystal structure of a mandelate racemase (muconate lactonizing enzyme2 family protein) from roseovarius nubinhibens
94 d1txga1
not modelled
10.8
25
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-likeSuperfamily: 6-phosphogluconate dehydrogenase C-terminal domain-likeFamily: Glycerol-3-phosphate dehydrogenase
95 c6o6dA_
not modelled
10.8
18
PDB header: ligaseChain: A: PDB Molecule: translation initiation factor if-3;PDBTitle: n-terminal domain of translation initiation factor if-3 from2 helicobacter pylori
96 c4v1ah_
not modelled
10.7
10
PDB header: ribosomeChain: H: PDB Molecule: PDBTitle: structure of the large subunit of the mammalian mitoribosome, part 22 of 2
97 c1jpdX_
not modelled
10.3
17
PDB header: isomeraseChain: X: PDB Molecule: l-ala-d/l-glu epimerase;PDBTitle: l-ala-d/l-glu epimerase
98 c3llhB_
not modelled
10.2
34
PDB header: rna binding proteinChain: B: PDB Molecule: risc-loading complex subunit tarbp2;PDBTitle: crystal structure of the first dsrbd of tar rna-binding protein 2
99 c2l2nA_
not modelled
10.2
15
PDB header: rna binding protein, plant proteinChain: A: PDB Molecule: hyponastic leave 1;PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for the first2 dsrbd of protein hyl1