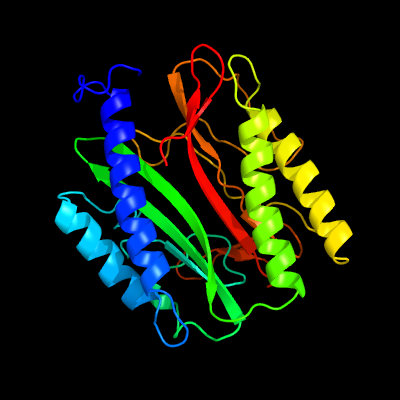
| 1 | c4fukB_
|
|
 |
100.0 |
35 |
PDB header:hydrolase
Chain: B: PDB Molecule:methionine aminopeptidase;
PDBTitle: aminopeptidase from trypanosoma brucei
|
|
|
|
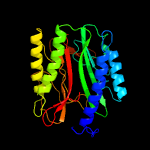
| 2 | c3tavA_
|
|
 |
100.0 |
74 |
PDB header:hydrolase
Chain: A: PDB Molecule:methionine aminopeptidase;
PDBTitle: crystal structure of a methionine aminopeptidase from mycobacterium2 abscessus
|
|
|
|
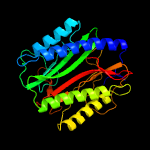
| 3 | c3s6bA_
|
|
 |
100.0 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:methionine aminopeptidase;
PDBTitle: crystal structure of methionine aminopeptidase 1b from plasmodium2 falciparum, pf10_0150
|
|
|
|
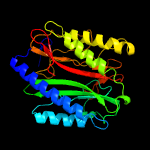
| 4 | c2g6pA_
|
|
 |
100.0 |
32 |
PDB header:hydrolase
Chain: A: PDB Molecule:methionine aminopeptidase 1;
PDBTitle: crystal structure of truncated (delta 1-89) human methionine2 aminopeptidase type 1 in complex with pyridyl pyrimidine derivative
|
|
|
|
| 5 | c2gz5A_
|
|
 |
100.0 |
32 |
PDB header:hydrolase
Chain: A: PDB Molecule:methionine aminopeptidase 1;
PDBTitle: human type 1 methionine aminopeptidase in complex with ovalicin at 1.12 ang
|
|
|
|
| 6 | c1yj3A_
|
|
 |
100.0 |
35 |
PDB header:hydrolase
Chain: A: PDB Molecule:methionine aminopeptidase;
PDBTitle: crystal structure analysis of product bound methionine aminopeptidase2 type 1c from mycobacterium tuberculosis
|
|
|
|
| 7 | d2gg2a1
|
|
 |
100.0 |
38 |
Fold:Creatinase/aminopeptidase
Superfamily:Creatinase/aminopeptidase
Family:Creatinase/aminopeptidase |
|
|
|
| 8 | d1o0xa_
|
|
 |
100.0 |
40 |
Fold:Creatinase/aminopeptidase
Superfamily:Creatinase/aminopeptidase
Family:Creatinase/aminopeptidase |
|
|
|
| 9 | c4fo7B_
|
|
 |
100.0 |
37 |
PDB header:hydrolase
Chain: B: PDB Molecule:methionine aminopeptidase;
PDBTitle: pseudomonas aeruginosa metap, in mn form
|
|
|
|
| 10 | c3mx6A_
|
|
 |
100.0 |
37 |
PDB header:hydrolase
Chain: A: PDB Molecule:methionine aminopeptidase;
PDBTitle: crystal structure of methionine aminopeptidase from rickettsia2 prowazekii bound to methionine
|
|
|
|
| 11 | c4km3B_
|
|
 |
100.0 |
36 |
PDB header:hydrolase
Chain: B: PDB Molecule:methionine aminopeptidase;
PDBTitle: discovery of a novel structural motif in methionine aminopeptidase2 from streptococci with possible post-translational modification
|
|
|
|
| 12 | c1w7vD_
|
|
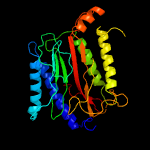 |
100.0 |
24 |
PDB header:hydrolase
Chain: D: PDB Molecule:xaa-pro aminopeptidase;
PDBTitle: znmg substituted aminopeptidase p from e. coli
|
|
|
|
| 13 | c4qr8B_
|
|
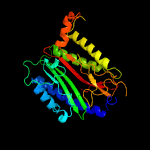 |
100.0 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:xaa-pro dipeptidase;
PDBTitle: crystal structure of e coli pepq
|
|
|
|
| 14 | c5wzeC_
|
|
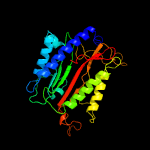 |
100.0 |
24 |
PDB header:hydrolase
Chain: C: PDB Molecule:aminopeptidase p;
PDBTitle: the structure of pseudomonas aeruginosa aminopeptidase pepp
|
|
|
|
| 15 | c3l24A_
|
|
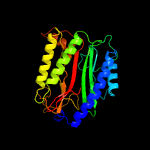 |
100.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:xaa-pro dipeptidase;
PDBTitle: crystal structure of the nerve agent degrading2 organophosphate anhydrolase/prolidase in complex with3 inhibitors
|
|
|
|
| 16 | d1qxya_
|
|
 |
100.0 |
32 |
Fold:Creatinase/aminopeptidase
Superfamily:Creatinase/aminopeptidase
Family:Creatinase/aminopeptidase |
|
|
|
| 17 | c3rvaA_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:organophosphorus acid anhydrolase;
PDBTitle: crystal structure of organophosphorus acid anhydrolase from2 alteromonas macleodii
|
|
|
|
| 18 | c5x49A_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable xaa-pro aminopeptidase 3;
PDBTitle: crystal structure of human mitochondrial x-prolyl aminopeptidase2 (xpnpep3)
|
|
|
|
| 19 | c2oknB_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:xaa-pro dipeptidase;
PDBTitle: crystal strcture of human prolidase
|
|
|
|
| 20 | c4r60A_
|
|
 |
100.0 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:proline dipeptidase;
PDBTitle: crystal structure of xaa-pro dipeptidase from xanthomonas campestris
|
|
|
|
| 21 | c6mrfA_ |
|
not modelled |
100.0 |
37 |
PDB header:hydrolase
Chain: A: PDB Molecule:methionine aminopeptidase;
PDBTitle: crystal structure of a methionine aminopeptidase metap from2 acinetobacter baumannii
|
|
|
| 22 | c3ig4E_ |
|
not modelled |
100.0 |
20 |
PDB header:hydrolase
Chain: E: PDB Molecule:xaa-pro aminopeptidase;
PDBTitle: structure of a putative aminopeptidase p from bacillus anthracis
|
|
|
| 23 | c1chmA_ |
|
not modelled |
100.0 |
19 |
PDB header:creatinase
Chain: A: PDB Molecule:creatine amidinohydrolase;
PDBTitle: enzymatic mechanism of creatine amidinohydrolase as deduced2 from crystal structures
|
|
|
| 24 | c2zsgB_ |
|
not modelled |
100.0 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:aminopeptidase p, putative;
PDBTitle: crystal structure of x-pro aminopeptidase from thermotoga maritima2 msb8
|
|
|
| 25 | c6a9vA_ |
|
not modelled |
100.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:intermediate cleaving peptidase 55;
PDBTitle: crystal structure of icp55 from saccharomyces cerevisiae (n-terminal2 42 residues deletion)
|
|
|
| 26 | c5cnxB_ |
|
not modelled |
100.0 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:aminopeptidase ypdf;
PDBTitle: crystal structure of xaa-pro aminopeptidase from escherichia coli k12
|
|
|
| 27 | c4egeA_ |
|
not modelled |
100.0 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:dipeptidase pepe;
PDBTitle: crystal structure of dipeptidase pepe from mycobacterium ulcerans
|
|
|
| 28 | c5ce6A_ |
|
not modelled |
100.0 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:fact-spt16;
PDBTitle: n-terminal domain of fact complex subunit spt16 from cicer arietinum2 (chickpea)
|
|
|
| 29 | d1b6aa2 |
|
not modelled |
100.0 |
22 |
Fold:Creatinase/aminopeptidase
Superfamily:Creatinase/aminopeptidase
Family:Creatinase/aminopeptidase |
|
|
| 30 | c5cdlA_ |
|
not modelled |
100.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:proline dipeptidase;
PDBTitle: proline dipeptidase from deinococcus radiodurans (selenomethionine2 derivative)
|
|
|
| 31 | c1wy2B_ |
|
not modelled |
100.0 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:xaa-pro dipeptidase;
PDBTitle: crystal structure of the prolidase from pyrococcus horikoshii ot3
|
|
|
| 32 | c3q6dA_ |
|
not modelled |
100.0 |
24 |
PDB header:viral protein
Chain: A: PDB Molecule:proline dipeptidase;
PDBTitle: xaa-pro dipeptidase from bacillus anthracis.
|
|
|
| 33 | c4fkcA_ |
|
not modelled |
100.0 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:xaa-pro aminopeptidase;
PDBTitle: recombinant prolidase from thermococcus sibiricus
|
|
|
| 34 | c2howB_ |
|
not modelled |
100.0 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:356aa long hypothetical dipeptidase;
PDBTitle: dipeptidase (ph0974) from pyrococcus horikoshii ot3
|
|
|
| 35 | c4zngA_ |
|
not modelled |
100.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:prolidase;
PDBTitle: x-ray crystallography of recombinant lactococcus lactis prolidase
|
|
|
| 36 | c3cb5A_ |
|
not modelled |
100.0 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:fact complex subunit spt16;
PDBTitle: crystal structure of the s. pombe peptidase homology domain of fact2 complex subunit spt16 (form a)
|
|
|
| 37 | d1chma2 |
|
not modelled |
100.0 |
19 |
Fold:Creatinase/aminopeptidase
Superfamily:Creatinase/aminopeptidase
Family:Creatinase/aminopeptidase |
|
|
| 38 | d2v3za2 |
|
not modelled |
100.0 |
24 |
Fold:Creatinase/aminopeptidase
Superfamily:Creatinase/aminopeptidase
Family:Creatinase/aminopeptidase |
|
|
| 39 | c5e5bA_ |
|
not modelled |
100.0 |
16 |
PDB header:transcription
Chain: A: PDB Molecule:fact complex subunit spt16;
PDBTitle: crystal structure of human spt16 n-terminal domain
|
|
|
| 40 | c5xevA_ |
|
not modelled |
100.0 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:xaa-pro dipeptidase,peptidase-related protein;
PDBTitle: crystal structure of a novel xaa-pro dipeptidase from deinococcus2 radiodurans
|
|
|
| 41 | d1pv9a2 |
|
not modelled |
100.0 |
22 |
Fold:Creatinase/aminopeptidase
Superfamily:Creatinase/aminopeptidase
Family:Creatinase/aminopeptidase |
|
|
| 42 | c6a8mA_ |
|
not modelled |
100.0 |
13 |
PDB header:dna binding protein
Chain: A: PDB Molecule:fact complex subunit spt16;
PDBTitle: n-terminal domain of fact complex subunit spt16 from eremothecium2 gossypii (ashbya gossypii)
|
|
|
| 43 | c3ctzA_ |
|
not modelled |
100.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:xaa-pro aminopeptidase 1;
PDBTitle: structure of human cytosolic x-prolyl aminopeptidase
|
|
|
| 44 | c4s2tP_ |
|
not modelled |
100.0 |
18 |
PDB header:hydrolase/hydrolase inhibitor
Chain: P: PDB Molecule:protein app-1;
PDBTitle: crystal structure of x-prolyl aminopeptidase from caenorhabditis2 elegans: a cytosolic enzyme with a di-nuclear active site
|
|
|
| 45 | c2v6cA_ |
|
not modelled |
100.0 |
23 |
PDB header:transcription regulator
Chain: A: PDB Molecule:proliferation-associated protein 2g4;
PDBTitle: crystal structure of erbb3 binding protein 1 (ebp1)
|
|
|
| 46 | c4b28A_ |
|
not modelled |
100.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:metallopeptidase, family m24, putative;
PDBTitle: crystal structure of dmsp lyase rddddp from roseobacter denitrificans
|
|
|
| 47 | c2q8kA_ |
|
not modelled |
100.0 |
23 |
PDB header:transcription
Chain: A: PDB Molecule:proliferation-associated protein 2g4;
PDBTitle: the crystal structure of ebp1
|
|
|
| 48 | c5jqkA_ |
|
not modelled |
100.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidase, putative;
PDBTitle: the xray crystal structure of p. falciparum aminopeptidase p
|
|
|
| 49 | d1xgsa2 |
|
not modelled |
100.0 |
31 |
Fold:Creatinase/aminopeptidase
Superfamily:Creatinase/aminopeptidase
Family:Creatinase/aminopeptidase |
|
|
| 50 | c3bitA_ |
|
not modelled |
100.0 |
12 |
PDB header:transcription
Chain: A: PDB Molecule:fact complex subunit spt16;
PDBTitle: crystal structure of yeast spt16 n-terminal domain
|
|
|
| 51 | c5jr6B_ |
|
not modelled |
100.0 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:peptidase, putative;
PDBTitle: the xray crystal structure of p. falciparum aminopeptidase p in2 complex with apstatin
|
|
|
| 52 | c1xgnB_ |
|
not modelled |
100.0 |
31 |
PDB header:aminopeptidase
Chain: B: PDB Molecule:methionine aminopeptidase;
PDBTitle: methionine aminopeptidase from hyperthermophile pyrococcus2 furiosus
|
|
|
| 53 | c1b6aA_ |
|
not modelled |
100.0 |
22 |
PDB header:angiogenesis inhibitor
Chain: A: PDB Molecule:methionine aminopeptidase;
PDBTitle: human methionine aminopeptidase 2 complexed with tnp-470
|
|
|
| 54 | c3fm3B_ |
|
not modelled |
100.0 |
25 |
PDB header:hydrolase
Chain: B: PDB Molecule:methionine aminopeptidase 2;
PDBTitle: crystal structure of an encephalitozoon cuniculi methionine2 aminopeptidase type 2
|
|
|
| 55 | c4ipaC_ |
|
not modelled |
100.0 |
20 |
PDB header:hydrolase
Chain: C: PDB Molecule:putative curved dna-binding protein;
PDBTitle: structure of a thermophilic arx1
|
|
|
| 56 | c1yw7A_ |
|
not modelled |
100.0 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:methionine aminopeptidase 2;
PDBTitle: h-metap2 complexed with a444148
|
|
|
| 57 | c3tb5C_ |
|
not modelled |
100.0 |
35 |
PDB header:hydrolase
Chain: C: PDB Molecule:methionine aminopeptidase;
PDBTitle: crystal structure of the enterococcus faecalis methionine2 aminopeptidase apo form
|
|
|
| 58 | c1kp0B_ |
|
not modelled |
100.0 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:creatine amidinohydrolase;
PDBTitle: the crystal structure analysis of creatine amidinohydrolase2 from actinobacillus
|
|
|
| 59 | d1kp0a2 |
|
not modelled |
100.0 |
17 |
Fold:Creatinase/aminopeptidase
Superfamily:Creatinase/aminopeptidase
Family:Creatinase/aminopeptidase |
|
|
| 60 | c4b6at_ |
|
not modelled |
100.0 |
17 |
PDB header:ribosome
Chain: T: PDB Molecule:60s ribosomal protein l21-a;
PDBTitle: cryo-em structure of the 60s ribosomal subunit in complex2 with arx1 and rei1
|
|
|
| 61 | c2y0oA_ |
|
not modelled |
28.6 |
27 |
PDB header:isomerase
Chain: A: PDB Molecule:probable d-lyxose ketol-isomerase;
PDBTitle: the structure of a d-lyxose isomerase from the sigmab2 regulon of bacillus subtilis
|
|
|
| 62 | c3mpbA_ |
|
not modelled |
27.2 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:sugar isomerase;
PDBTitle: z5688 from e. coli o157:h7 bound to fructose
|
|
|
| 63 | c4mozC_ |
|
not modelled |
24.8 |
24 |
PDB header:lyase
Chain: C: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: fructose-bisphosphate aldolase from slackia heliotrinireducens dsm2 20476
|
|
|
| 64 | c3jr1A_ |
|
not modelled |
21.2 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:putative fructosamine-3-kinase;
PDBTitle: crystal structure of putative fructosamine-3-kinase (yp_719053.1) from2 haemophilus somnus 129pt at 2.32 a resolution
|
|
|
| 65 | d1kt0a2 |
|
not modelled |
20.0 |
24 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
|
|
| 66 | d1j3ba1 |
|
not modelled |
19.9 |
14 |
Fold:PEP carboxykinase-like
Superfamily:PEP carboxykinase-like
Family:PEP carboxykinase C-terminal domain |
|
|
| 67 | c2hl7A_ |
|
not modelled |
19.1 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome c-type biogenesis protein ccmh;
PDBTitle: crystal structure of the periplasmic domain of ccmh from pseudomonas2 aeruginosa
|
|
|
| 68 | d1l1pa_ |
|
not modelled |
18.1 |
30 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
|
|
| 69 | c2kw0A_ |
|
not modelled |
18.0 |
6 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ccmh protein;
PDBTitle: solution structure of n-terminal domain of ccmh from escherichia.coli
|
|
|
| 70 | d1w26a3 |
|
not modelled |
16.7 |
16 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
|
|
| 71 | c2c5qE_ |
|
not modelled |
16.6 |
14 |
PDB header:structural genomics,unknown function
Chain: E: PDB Molecule:rraa-like protein yer010c;
PDBTitle: crystal structure of yeast yer010cp
|
|
|
| 72 | c3prdA_ |
|
not modelled |
16.1 |
24 |
PDB header:chaperone, isomerase
Chain: A: PDB Molecule:fkbp-type peptidyl-prolyl cis-trans isomerase;
PDBTitle: structural analysis of protein folding by the methanococcus jannaschii2 chaperone fkbp26
|
|
|
| 73 | c2ki0A_ |
|
not modelled |
14.4 |
23 |
PDB header:de novo protein
Chain: A: PDB Molecule:ds119;
PDBTitle: nmr structure of a de novo designed beta alpha beta
|
|
|
| 74 | c2yqkA_ |
|
not modelled |
13.9 |
13 |
PDB header:transcription/apoptosis
Chain: A: PDB Molecule:arginine-glutamic acid dipeptide repeats protein;
PDBTitle: solution structure of the sant domain in arginine-glutamic2 acid dipeptide (re) repeats
|
|
|
| 75 | d1t11a3 |
|
not modelled |
11.5 |
40 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
|
|
| 76 | c2k3uA_ |
|
not modelled |
10.5 |
31 |
PDB header:immune system
Chain: A: PDB Molecule:chemotaxis inhibitory protein;
PDBTitle: structure of the tyrosine-sulfated c5a receptor n-terminus2 in complex with the immune evasion protein chips.
|
|
|
| 77 | d1xc5a1 |
|
not modelled |
10.5 |
11 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Myb/SANT domain |
|
|
| 78 | d1iwpg_ |
|
not modelled |
10.2 |
26 |
Fold:Open three-helical up-and-down bundle
Superfamily:Diol dehydratase, gamma subunit
Family:Diol dehydratase, gamma subunit |
|
|
| 79 | c1uc5M_ |
|
not modelled |
10.0 |
23 |
PDB header:lyase
Chain: M: PDB Molecule:diol dehydrase gamma subunit;
PDBTitle: structure of diol dehydratase complexed with (r)-1,2-2 propanediol
|
|
|
| 80 | d1eexg_ |
|
not modelled |
10.0 |
23 |
Fold:Open three-helical up-and-down bundle
Superfamily:Diol dehydratase, gamma subunit
Family:Diol dehydratase, gamma subunit |
|
|
| 81 | c2pc9B_ |
|
not modelled |
9.8 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:phosphoenolpyruvate carboxykinase [atp];
PDBTitle: crystal structure of atp-dependent phosphoenolpyruvate carboxykinase2 from thermus thermophilus hb8
|
|
|
| 82 | c5i1tA_ |
|
not modelled |
9.1 |
44 |
PDB header:hydrolase
Chain: A: PDB Molecule:stage ii sporulation protein d;
PDBTitle: 2.6 angstrom resolution crystal structure of stage ii sporulation2 protein d (spoiid) from clostridium difficile in complex with3 triacetylchitotriose
|
|
|
| 83 | c4qblF_ |
|
not modelled |
8.9 |
15 |
PDB header:hydrolase
Chain: F: PDB Molecule:vrr-nuc;
PDBTitle: vrr_nuc domain protein
|
|
|
| 84 | c4rwrB_ |
|
not modelled |
8.9 |
28 |
PDB header:viral protein
Chain: B: PDB Molecule:stage ii sporulation protein d;
PDBTitle: 2.1 angstrom crystal structure of stage ii sporulation protein d from2 bacillus anthracis
|
|
|
| 85 | d2crga1 |
|
not modelled |
8.9 |
25 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Myb/SANT domain |
|
|
| 86 | c6fx6A_ |
|
not modelled |
8.0 |
38 |
PDB header:unknown function
Chain: A: PDB Molecule:satie-ted;
PDBTitle: thioester domain of the staphylococcus aureus tie protein
|
|
|
| 87 | c5zr0A_ |
|
not modelled |
7.7 |
27 |
PDB header:chaperone
Chain: A: PDB Molecule:maltose-binding periplasmic protein,trigger factor;
PDBTitle: solution structure of peptidyl-prolyl cis/trans isomerase domain of2 trigger factor in complex with mbp
|
|
|
| 88 | c1w26B_ |
|
not modelled |
7.6 |
16 |
PDB header:chaperone
Chain: B: PDB Molecule:trigger factor;
PDBTitle: trigger factor in complex with the ribosome forms a2 molecular cradle for nascent proteins
|
|
|
| 89 | c3zieC_ |
|
not modelled |
7.5 |
31 |
PDB header:cell cycle
Chain: C: PDB Molecule:sepf-like protein;
PDBTitle: sepf-like protein from archaeoglobus fulgidus
|
|
|
| 90 | c2kxxA_ |
|
not modelled |
7.3 |
5 |
PDB header:protein binding
Chain: A: PDB Molecule:small protein a;
PDBTitle: nmr structure of escherichia coli bame, a lipoprotein component of the2 beta-barrel assembly machinery complex
|
|
|
| 91 | d2k57a1 |
|
not modelled |
7.2 |
28 |
Fold:Sm-like fold
Superfamily:Sm-like ribonucleoproteins
Family:YgdI/YgdR-like |
|
|
| 92 | d1m1fa_ |
|
not modelled |
7.1 |
6 |
Fold:SH3-like barrel
Superfamily:Cell growth inhibitor/plasmid maintenance toxic component
Family:Kid/PemK |
|
|
| 93 | c2ls01_ |
|
not modelled |
7.0 |
19 |
PDB header:hydrolase
Chain: 1: PDB Molecule:zoocin a endopeptidase;
PDB Fragment:unp residues 170-285;
PDBTitle: solution structure of the target recognition domain of zoocin a
|
|
|
| 94 | c3nojA_ |
|
not modelled |
7.0 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:4-carboxy-4-hydroxy-2-oxoadipate aldolase/oxaloacetate
PDBTitle: the structure of hmg/cha aldolase from the protocatechuate degradation2 pathway of pseudomonas putida
|
|
|
| 95 | c4mspB_ |
|
not modelled |
6.9 |
29 |
PDB header:isomerase
Chain: B: PDB Molecule:peptidyl-prolyl cis-trans isomerase fkbp14;
PDBTitle: crystal structure of human peptidyl-prolyl cis-trans isomerase fkbp222 (aka fkbp14) containing two ef-hand motifs
|
|
|
| 96 | c4d4pG_ |
|
not modelled |
6.6 |
24 |
PDB header:translation
Chain: G: PDB Molecule:protein ats1, diphthamide biosynthesis protein 3;
PDBTitle: crystal structure of the kti11 kti13 heterodimer spacegroup p65
|
|
|
| 97 | c1kt0A_ |
|
not modelled |
6.5 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:51 kda fk506-binding protein;
PDBTitle: structure of the large fkbp-like protein, fkbp51, involved in steroid2 receptor complexes
|
|
|
| 98 | c1hxvA_ |
|
not modelled |
6.5 |
41 |
PDB header:chaperone
Chain: A: PDB Molecule:trigger factor;
PDBTitle: ppiase domain of the mycoplasma genitalium trigger factor
|
|
|
| 99 | d1hxva_ |
|
not modelled |
6.5 |
41 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
|
|