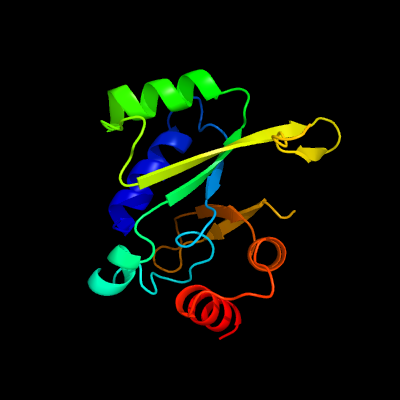
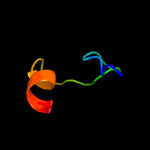
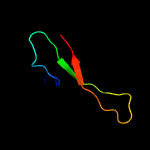
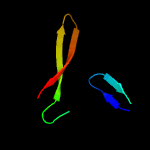
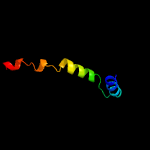
1 c4e8iA_
86.8
16
PDB header: transferaseChain: A: PDB Molecule: lincosamide resistance protein;PDBTitle: crystal structure of lincosamide antibiotic adenylyltransferase lina,2 apo
2 d1no5a_
54.7
24
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Catalytic subunit of bi-partite nucleotidyltransferase
3 d1wota_
54.2
37
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Catalytic subunit of bi-partite nucleotidyltransferase
4 c2mdwA_
50.3
50
PDB header: de novo proteinChain: A: PDB Molecule: designed protein;PDBTitle: nmr structure of a strand-swapped dimer of the ww domain
5 c2ky8A_
48.6
25
PDB header: transcription/dnaChain: A: PDB Molecule: methyl-cpg-binding domain protein 2;PDBTitle: solution structure and dynamic analysis of chicken mbd2 methyl binding2 domain bound to a target methylated dna sequence
6 c3fm2A_
44.1
32
PDB header: heme-binding proteinChain: A: PDB Molecule: uncharacterized protein, distantly related to a hemePDBTitle: crystal structure of a putative heme-binding protein (ava_4353) from2 anabaena variabilis atcc 29413 at 1.80 a resolution
7 d1ig4a_
39.4
22
Fold: DNA-binding domainSuperfamily: DNA-binding domainFamily: Methyl-CpG-binding domain, MBD
8 d1ylqa1
38.4
17
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Catalytic subunit of bi-partite nucleotidyltransferase
9 c2l9dA_
36.3
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of the protein yp_546394.1, the first structural2 representative of the pfam family pf12112
10 d1ub1a_
35.8
25
Fold: DNA-binding domainSuperfamily: DNA-binding domainFamily: Methyl-CpG-binding domain, MBD
11 c3zvrA_
33.7
23
PDB header: hydrolaseChain: A: PDB Molecule: dynamin-1;PDBTitle: crystal structure of dynamin
12 d1tu1a_
30.6
19
Fold: Mog1p/PsbP-likeSuperfamily: Mog1p/PsbP-likeFamily: PA0094-like
13 d2gc6a1
27.8
17
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: Folylpolyglutamate synthetase, C-terminal domain
14 c3snhA_
26.3
44
PDB header: endocytosisChain: A: PDB Molecule: dynamin-1;PDBTitle: crystal structure of nucleotide-free human dynamin1
15 c5xmcA_
25.3
32
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase itchy;PDBTitle: crystal structure of the auto-inhibited nedd4 family e3 ligase itch
16 d2qdya1
23.0
27
Fold: Nitrile hydratase alpha chainSuperfamily: Nitrile hydratase alpha chainFamily: Nitrile hydratase alpha chain
17 d1v29a_
22.9
27
Fold: Nitrile hydratase alpha chainSuperfamily: Nitrile hydratase alpha chainFamily: Nitrile hydratase alpha chain
18 c3qyhG_
22.6
31
PDB header: lyaseChain: G: PDB Molecule: co-type nitrile hydratase alpha subunit;PDBTitle: crystal structure of co-type nitrile hydratase beta-h71l from2 pseudomonas putida.
19 c4l9cA_
22.5
20
PDB header: protein bindingChain: A: PDB Molecule: f-box only protein 7;PDBTitle: crystal structure of the fp domain of human f-box protein fbxo72 (native)
20 c2yscA_
22.4
38
PDB header: protein bindingChain: A: PDB Molecule: amyloid beta a4 precursor protein-binding familyPDBTitle: solution structure of the ww domain from the human amyloid2 beta a4 precursor protein-binding family b member 3, apbb3
21 d1qk9a_
not modelled
21.7
26
Fold: DNA-binding domainSuperfamily: DNA-binding domainFamily: Methyl-CpG-binding domain, MBD
22 c3kepA_
not modelled
21.0
11
PDB header: protein transport, rna binding proteinChain: A: PDB Molecule: nucleoporin nup145;PDBTitle: crystal structure of the autoproteolytic domain from the nuclear pore2 complex component nup145 from saccharomyces cerevisiae
23 c2kq0A_
not modelled
20.4
56
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase nedd4;PDBTitle: human nedd4 3rd ww domain complex with ebola zaire virus matrix2 protein vp40 derived peptide ilptappeymea
24 c2z0bB_
not modelled
20.0
27
PDB header: hydrolaseChain: B: PDB Molecule: putative glycerophosphodiester phosphodiesterase 5;PDBTitle: crystal structure of cbm20 domain of human putative2 glycerophosphodiester phosphodiesterase 5 (kiaa1434)
25 c4yntA_
not modelled
19.3
27
PDB header: oxidoreductaseChain: A: PDB Molecule: glucose oxidase, putative;PDBTitle: crystal structure of aspergillus flavus fad glucose dehydrogenase
26 c1ju2A_
not modelled
18.9
30
PDB header: lyaseChain: A: PDB Molecule: hydroxynitrile lyase;PDBTitle: crystal structure of the hydroxynitrile lyase from almond
27 c1naaB_
not modelled
18.2
22
PDB header: oxidoreductaseChain: B: PDB Molecule: cellobiose dehydrogenase;PDBTitle: cellobiose dehydrogenase flavoprotein fragment in complex with2 cellobionolactam
28 c2lazA_
not modelled
18.2
44
PDB header: signaling protein/transcriptionChain: A: PDB Molecule: e3 ubiquitin-protein ligase smurf1;PDBTitle: structure of the first ww domain of human smurf1 in complex with a2 mono-phosphorylated human smad1 derived peptide
29 c2lb0A_
not modelled
18.2
44
PDB header: signaling protein/transcriptionChain: A: PDB Molecule: e3 ubiquitin-protein ligase smurf1;PDBTitle: structure of the first ww domain of human smurf1 in complex with a di-2 phosphorylated human smad1 derived peptide
30 c1gpeA_
not modelled
18.1
20
PDB header: oxidoreductase(flavoprotein)Chain: A: PDB Molecule: protein (glucose oxidase);PDBTitle: glucose oxidase from penicillium amagasakiense
31 c6ql4B_
not modelled
17.9
56
PDB header: motor proteinChain: B: PDB Molecule: putative mitochondrial dynamin protein;PDBTitle: crystal structure of nucleotide-free mgm1
32 c3vxvA_
not modelled
17.6
19
PDB header: hydrolase/dnaChain: A: PDB Molecule: methyl-cpg-binding domain protein 4;PDBTitle: crystal structure of methyl cpg binding domain of mbd4 in complex with2 the 5mcg/tg sequence
33 d1kdga1
not modelled
17.6
22
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
34 d1ugpa_
not modelled
17.5
27
Fold: Nitrile hydratase alpha chainSuperfamily: Nitrile hydratase alpha chainFamily: Nitrile hydratase alpha chain
35 d1cgta2
not modelled
17.0
12
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: Starch-binding domain
36 c5nccB_
not modelled
16.9
20
PDB header: oxidoreductaseChain: B: PDB Molecule: fatty acid photodecarboxylase;PDBTitle: structure of fatty acid photodecarboxylase in complex with fad and2 palmitic acid
37 d1cf3a1
not modelled
16.8
30
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
38 d2itka1
not modelled
16.8
44
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain
39 c3q9tB_
not modelled
16.6
20
PDB header: oxidoreductaseChain: B: PDB Molecule: choline dehydrogenase and related flavoproteins;PDBTitle: crystal structure analysis of formate oxidase
40 c1wr4A_
not modelled
16.6
38
PDB header: ligaseChain: A: PDB Molecule: ubiquitin-protein ligase nedd4-2;PDBTitle: solution structure of the second ww domain of nedd4-2
41 c3fimB_
not modelled
16.6
44
PDB header: oxidoreductaseChain: B: PDB Molecule: aryl-alcohol oxidase;PDBTitle: crystal structure of aryl-alcohol-oxidase from pleurotus eryingii
42 c5aa6F_
not modelled
16.6
38
PDB header: oxidoreductaseChain: F: PDB Molecule: vanadium-dependent bromoperoxidase 2;PDBTitle: homohexameric structure of the second vanadate-dependent2 bromoperoxidase (anii) from ascophyllum nodosum
43 d1k9ra_
not modelled
16.3
36
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain
44 c4h7uA_
not modelled
16.2
44
PDB header: oxidoreductaseChain: A: PDB Molecule: pyranose dehydrogenase;PDBTitle: crystal structure of pyranose dehydrogenase from agaricus meleagris,2 wildtype
45 c2n8tA_
not modelled
16.0
31
PDB header: ligase/peptideChain: A: PDB Molecule: e3 ubiquitin-protein ligase nedd4;PDBTitle: solution structure of the rnedd4 ww2 domain-cx43ct peptide complex by2 nmr
46 c4qi4A_
not modelled
15.7
33
PDB header: oxidoreductaseChain: A: PDB Molecule: cellobiose dehydrogenase;PDBTitle: dehydrogenase domain of myriococcum thermophilum cellobiose2 dehydrogenase, mtdh
47 c2jmfA_
not modelled
15.4
38
PDB header: ligase/signaling proteinChain: A: PDB Molecule: e3 ubiquitin-protein ligase suppressor of deltex;PDBTitle: solution structure of the su(dx) ww4- notch py peptide2 complex
48 c4fm4C_
not modelled
15.4
18
PDB header: lyaseChain: C: PDB Molecule: nitrile hydratase alpha subunit;PDBTitle: wild type fe-type nitrile hydratase from comamonas testosteroni ni1
49 d2f21a1
not modelled
15.3
56
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain
50 c3pz8A_
not modelled
15.3
44
PDB header: signaling proteinChain: A: PDB Molecule: segment polarity protein dishevelled homolog dvl-1;PDBTitle: crystal structure of dvl1-dix(y17d) mutant
51 d1pina1
not modelled
15.3
56
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain
52 c1cf3A_
not modelled
14.9
30
PDB header: oxidoreductase(flavoprotein)Chain: A: PDB Molecule: protein (glucose oxidase);PDBTitle: glucose oxidase from apergillus niger
53 c3qw4B_
not modelled
14.8
19
PDB header: transferase, lyaseChain: B: PDB Molecule: ump synthase;PDBTitle: structure of leishmania donovani ump synthase
54 c6h3gC_
not modelled
14.5
50
PDB header: oxidoreductaseChain: C: PDB Molecule: alcohol oxidase;PDBTitle: alcohol oxidase from phanerochaete chrysosporium
55 d1cyga2
not modelled
14.5
25
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: Starch-binding domain
56 c2yshA_
not modelled
14.4
29
PDB header: protein bindingChain: A: PDB Molecule: growth-arrest-specific protein 7;PDBTitle: solution structure of the ww domain from the human growth-2 arrest-specific protein 7, gas-7
57 c4whjA_
not modelled
14.3
44
PDB header: antiviral protein, hydrolaseChain: A: PDB Molecule: interferon-induced gtp-binding protein mx2;PDBTitle: myxovirus resistance protein 2 (mxb)
58 c5hsaG_
not modelled
14.3
42
PDB header: oxidoreductaseChain: G: PDB Molecule: alcohol oxidase 1;PDBTitle: alcohol oxidase aox1 from pichia pastoris
59 c4n0iB_
not modelled
14.2
24
PDB header: ligaseChain: B: PDB Molecule: glutamyl-trna(gln) amidotransferase subunit b,PDBTitle: crystal structure of s. cerevisiae mitochondrial gatfab in complex2 with glutamine
60 c2jbvA_
not modelled
14.1
30
PDB header: oxidoreductaseChain: A: PDB Molecule: choline oxidase;PDBTitle: crystal structure of choline oxidase reveals insights into the2 catalytic mechanism
61 c3ip4B_
not modelled
13.9
30
PDB header: ligaseChain: B: PDB Molecule: aspartyl/glutamyl-trna(asn/gln) amidotransferase subunit b;PDBTitle: the high resolution structure of gatcab
62 c2ebuA_
not modelled
13.9
28
PDB header: replicationChain: A: PDB Molecule: replication factor c subunit 1;PDBTitle: solution structure of the brct domain from human2 replication factor c large subunit 1
63 d1f8ab1
not modelled
13.8
43
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain
64 c2reeB_
not modelled
13.7
18
PDB header: transferase, lyaseChain: B: PDB Molecule: cura;PDBTitle: crystal structure of the loading gnatl domain of cura from lyngbya2 majuscula
65 d2f5va1
not modelled
13.6
44
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
66 d1prtb1
not modelled
13.6
29
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits
67 c1e0mA_
not modelled
13.4
44
PDB header: de novo proteinChain: A: PDB Molecule: wwprototype;PDBTitle: prototype ww domain
68 d1i8gb_
not modelled
13.4
56
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain
69 d1cxla2
not modelled
13.4
47
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: Starch-binding domain
70 c3t37A_
not modelled
13.2
22
PDB header: oxidoreductaseChain: A: PDB Molecule: probable dehydrogenase;PDBTitle: crystal structure of pyridoxine 4-oxidase from mesorbium loti
71 d1cw0a_
not modelled
13.1
18
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Very short patch repair (VSR) endonuclease
72 d2f2ab2
not modelled
13.0
30
Fold: Glutamine synthetase/guanido kinaseSuperfamily: Glutamine synthetase/guanido kinaseFamily: GatB/GatE catalytic domain-like
73 c4udpA_
not modelled
13.0
25
PDB header: oxidoreductaseChain: A: PDB Molecule: glucose-methanol-choline oxidoreductase;PDBTitle: crystal structure of 5-hydroxymethylfurfural oxidase (hmfo) in the2 oxidized state
74 d1ju2a1
not modelled
12.8
22
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
75 c2l4jA_
not modelled
12.8
25
PDB header: transcriptionChain: A: PDB Molecule: yes-associated protein 2 (yap2);PDBTitle: yap ww2
76 d2jmfa1
not modelled
12.7
63
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain
77 d1y7pa1
not modelled
12.6
28
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: AF1403 C-terminal domain-like
78 c1coyA_
not modelled
12.6
50
PDB header: oxidoreductase(oxygen receptor)Chain: A: PDB Molecule: cholesterol oxidase;PDBTitle: crystal structure of cholesterol oxidase complexed with a steroid2 substrate. implications for fad dependent alcohol oxidases
79 d1gpea1
not modelled
12.5
20
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
80 c3al0B_
not modelled
12.3
36
PDB header: ligase/rnaChain: B: PDB Molecule: aspartyl/glutamyl-trna(asn/gln) amidotransferase subunit b;PDBTitle: crystal structure of the glutamine transamidosome from thermotoga2 maritima in the glutamylation state.
81 c2ysgA_
not modelled
12.3
36
PDB header: protein bindingChain: A: PDB Molecule: syntaxin-binding protein 4;PDBTitle: solution structure of the ww domain from the human syntaxin-2 binding protein 4
82 c2g5iB_
not modelled
12.2
30
PDB header: ligaseChain: B: PDB Molecule: aspartyl/glutamyl-trna(asn/gln) amidotransferasePDBTitle: structure of trna-dependent amidotransferase gatcab2 complexed with adp-alf4
83 c1s3bB_
not modelled
12.2
40
PDB header: oxidoreductaseChain: B: PDB Molecule: amine oxidase [flavin-containing] b;PDBTitle: crystal structure of maob in complex with n-methyl-n-2 propargyl-1(r)-aminoindan
84 c1yw5A_
not modelled
12.2
25
PDB header: isomeraseChain: A: PDB Molecule: peptidyl prolyl cis/trans isomerase;PDBTitle: peptidyl-prolyl isomerase ess1 from candida albicans
85 c2dmvA_
not modelled
12.2
38
PDB header: ligaseChain: A: PDB Molecule: itchy homolog e3 ubiquitin protein ligase;PDBTitle: solution structure of the second ww domain of itchy homolog2 e3 ubiquitin protein ligase (itch)
86 d1tk7a2
not modelled
12.1
56
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain
87 c1ymzA_
not modelled
11.9
56
PDB header: unknown functionChain: A: PDB Molecule: cc45;PDBTitle: cc45, an artificial ww domain designed using statistical2 coupling analysis
88 d1d7ya3
not modelled
11.8
19
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domainFamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domain
89 c2ysfA_
not modelled
11.8
56
PDB header: protein bindingChain: A: PDB Molecule: e3 ubiquitin-protein ligase itchy homolog;PDBTitle: solution structure of the fourth ww domain from the human2 e3 ubiquitin-protein ligase itchy homolog, itch
90 c2ez5W_
not modelled
11.7
23
PDB header: signalling protein,ligaseChain: W: PDB Molecule: e3 ubiquitin-protein ligase nedd4;PDBTitle: solution structure of the dnedd4 ww3* domain- comm lpsy2 peptide complex
91 c4p4sA_
not modelled
11.6
44
PDB header: antiviral protein/hydrolaseChain: A: PDB Molecule: interferon-induced gtp-binding protein mx1;PDBTitle: gmppcp-bound stalkless-mxa
92 c5b3zB_
not modelled
11.4
50
PDB header: isomerase,sugar binding proteinChain: B: PDB Molecule: peptidyl-prolyl cis-trans isomerase nima-interacting 1,PDBTitle: crystal structure of hpin1 ww domain (5-39) fused with maltose-binding2 protein
93 d1qhoa2
not modelled
11.4
20
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: Starch-binding domain
94 d1tk7a1
not modelled
11.3
31
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain
95 c1wr7A_
not modelled
11.3
56
PDB header: ligaseChain: A: PDB Molecule: nedd4-2;PDBTitle: solution structure of the third ww domain of nedd4-2
96 d3bmva2
not modelled
11.3
33
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: Starch-binding domain
97 c2gewA_
not modelled
11.1
40
PDB header: oxidoreductaseChain: A: PDB Molecule: cholesterol oxidase;PDBTitle: atomic resolution structure of cholesterol oxidase @ ph 9.02 (streptomyces sp. sa-coo)
98 d1nmva1
not modelled
11.1
43
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain
99 c1dxlC_
not modelled
11.0
27
PDB header: oxidoreductaseChain: C: PDB Molecule: dihydrolipoamide dehydrogenase;PDBTitle: dihydrolipoamide dehydrogenase of glycine decarboxylase2 from pisum sativum