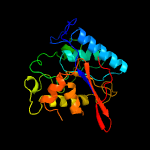
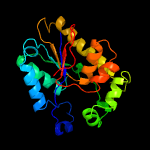
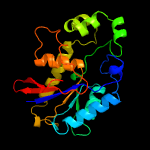
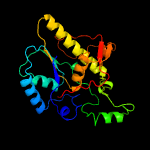
1 c4pz9B_
100.0
20
PDB header: hydrolaseChain: B: PDB Molecule: glucosyl-3-phosphoglycerate phosphatase;PDBTitle: the native structure of mycobacterial glucosyl-3-phosphoglycerate2 phosphatase rv2419c
2 c3f3kA_
100.0
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ykr043c;PDBTitle: the structure of uncharacterized protein ykr043c from saccharomyces2 cerevisiae.
3 c2ikqA_
99.9
20
PDB header: signaling protein, immune systemChain: A: PDB Molecule: suppressor of t-cell receptor signaling 1;PDBTitle: crystal structure of mouse sts-1 pgm domain in complex with phosphate
4 c3r7aA_
99.9
15
PDB header: transferaseChain: A: PDB Molecule: phosphoglycerate mutase, putative;PDBTitle: crystal structure of phosphoglycerate mutase from bacillus anthracis2 str. sterne
5 c2yn0A_
99.9
14
PDB header: transcriptionChain: A: PDB Molecule: transcription factor tau 55 kda subunit;PDBTitle: tau55 histidine phosphatase domain
6 c4ij5B_
99.9
16
PDB header: hydrolaseChain: B: PDB Molecule: phosphoserine phosphatase 1;PDBTitle: crystal structure of a novel-type phosphoserine phosphatase from2 hydrogenobacter thermophilus tk-6
7 c3d4iD_
99.9
23
PDB header: hydrolaseChain: D: PDB Molecule: sts-2 protein;PDBTitle: crystal structure of the 2h-phosphatase domain of sts-2
8 c2qniA_
99.9
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu0299;PDBTitle: crystal structure of uncharacterized protein atu0299
9 d1h2ea_
99.9
18
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase
10 c3ll4B_
99.9
16
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized protein ykr043c;PDBTitle: structure of the h13a mutant of ykr043c in complex with fructose-1,6-2 bisphosphate
11 d1e58a_
99.9
17
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase
12 c5zkkA_
99.9
20
PDB header: hydrolaseChain: A: PDB Molecule: phosphoglycerate mutase family protein, putative;PDBTitle: crystal structure of phosphoserine phosphatase from entamoeba2 histolytica
13 c4embD_
99.9
18
PDB header: isomeraseChain: D: PDB Molecule: 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase;PDBTitle: crystal structure of a phosphoglycerate mutase gpma from borrelia2 burgdorferi b31
14 c3e9eB_
99.9
20
PDB header: hydrolaseChain: B: PDB Molecule: zgc:56074;PDBTitle: structure of full-length h11a mutant form of tigar from danio rerio
15 d1riia_
99.9
19
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase
16 c1yjxD_
99.9
16
PDB header: isomerase, hydrolaseChain: D: PDB Molecule: phosphoglycerate mutase 1;PDBTitle: crystal structure of human b type phosphoglycerate mutase
17 c3dcyA_
99.9
19
PDB header: apoptosis regulatorChain: A: PDB Molecule: regulator protein;PDBTitle: crystal structure a tp53-induced glycolysis and apoptosis regulator2 protein from homo sapiens.
18 c5vveA_
99.9
17
PDB header: isomeraseChain: A: PDB Molecule: phosphoglycerate mutase;PDBTitle: crystal structure of phosphoglycerate mutase from naegleria fowleri
19 c2yn2A_
99.9
14
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein ynl108c;PDBTitle: huf protein - paralogue of the tau55 histidine phosphatase domain
20 d1xq9a_
99.9
19
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase
21 c2i1vB_
not modelled
99.9
19
PDB header: transferase, hydrolaseChain: B: PDB Molecule: 6-phosphofructo-2-kinase/fructose-2,6-PDBTitle: crystal structure of pfkfb3 in complex with adp and2 fructose-2,6-bisphosphate
22 c3c7tB_
not modelled
99.9
23
PDB header: hydrolaseChain: B: PDB Molecule: ecdysteroid-phosphate phosphatase;PDBTitle: crystal structure of the ecdysone phosphate phosphatase, eppase, from2 bombix mori in complex with tungstate
23 d1fzta_
not modelled
99.9
16
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase
24 c3eznB_
not modelled
99.9
16
PDB header: isomeraseChain: B: PDB Molecule: 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase;PDBTitle: crystal structure of phosphoglyceromutase from burkholderia2 pseudomallei 1710b
25 d1bifa2
not modelled
99.9
19
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, phosphatase domain
26 c1k6mA_
not modelled
99.9
22
PDB header: transferase, hydrolaseChain: A: PDB Molecule: 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2-PDBTitle: crystal structure of human liver 6-phosphofructo-2-kinase/fructose-2,2 6-bisphosphatase
27 c3hjgB_
not modelled
99.9
21
PDB header: hydrolaseChain: B: PDB Molecule: putative alpha-ribazole-5'-phosphate phosphatase cobc;PDBTitle: crystal structure of putative alpha-ribazole-5'-phosphate phosphatase2 cobc from vibrio parahaemolyticus
28 d1qhfa_
not modelled
99.9
19
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase
29 d2hhja1
not modelled
99.9
18
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase
30 c2a6pA_
not modelled
99.9
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: possible phosphoglycerate mutase gpm2;PDBTitle: structure solution to 2.2 angstrom and functional characterisation of2 the open reading frame rv3214 from mycobacterium tuberculosis
31 c1bifA_
not modelled
99.9
20
PDB header: bifunctional enzymeChain: A: PDB Molecule: 6-phosphofructo-2-kinase/ fructose-2,6-bisphosphatase;PDBTitle: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase bifunctional2 enzyme complexed with atp-g-s and phosphate
32 c3d8hB_
not modelled
99.9
19
PDB header: isomeraseChain: B: PDB Molecule: glycolytic phosphoglycerate mutase;PDBTitle: crystal structure of phosphoglycerate mutase from cryptosporidium2 parvum, cgd7_4270
33 c4eo9A_
not modelled
99.9
19
PDB header: isomeraseChain: A: PDB Molecule: 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase;PDBTitle: crystal structure of a phosphoglycerate mutase gpm1 from mycobacterium2 leprae
34 d3pgma_
not modelled
99.9
20
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase
35 c5um0A_
not modelled
99.9
18
PDB header: isomeraseChain: A: PDB Molecule: 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase;PDBTitle: crystal structure of 2,3-bisphosphoglycerate-dependent2 phosphoglycerate mutase from neisseria gonorrhoeae
36 c3mxoB_
not modelled
99.9
21
PDB header: hydrolaseChain: B: PDB Molecule: serine/threonine-protein phosphatase pgam5, mitochondrial;PDBTitle: crystal structure oh human phosphoglycerate mutase family member 52 (pgam5)
37 c6e4bC_
not modelled
99.9
20
PDB header: hydrolaseChain: C: PDB Molecule: adenosylcobalamin/alpha-ribazole phosphatase;PDBTitle: the crystal structure of a putative alpha-ribazole-5'-p phosphatase2 from escherichia coli str. k-12 substr. mg1655
38 d1k6ma2
not modelled
99.9
22
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, phosphatase domain
39 d1tipa_
not modelled
99.9
20
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, phosphatase domain
40 c2rflB_
not modelled
99.8
19
PDB header: hydrolase, isomeraseChain: B: PDB Molecule: putative phosphohistidine phosphatase sixa;PDBTitle: crystal structure of the putative phosphohistidine phosphatase sixa2 from agrobacterium tumefaciens
41 c3eozB_
not modelled
99.8
20
PDB header: isomeraseChain: B: PDB Molecule: putative phosphoglycerate mutase;PDBTitle: crystal structure of phosphoglycerate mutase from plasmodium2 falciparum, pfd0660w
42 d1v37a_
not modelled
99.8
23
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Cofactor-dependent phosphoglycerate mutase
43 c1ujcA_
not modelled
99.8
21
PDB header: hydrolaseChain: A: PDB Molecule: phosphohistidine phosphatase sixa;PDBTitle: structure of the protein histidine phosphatase sixa2 complexed with tungstate
44 c5gg7A_
not modelled
99.7
18
PDB header: hydrolaseChain: A: PDB Molecule: hydrolase, nudix family protein;PDBTitle: crystal structure of mycobacterium smegmatis mutt1 in complex with 8-2 oxo-dgtp, 8-oxo-dgmp and pyrophosphate (i)
45 c3f2iD_
not modelled
99.6
19
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: alr0221 protein;PDBTitle: crystal structure of the alr0221 protein from nostoc, northeast2 structural genomics consortium target nsr422.
46 c3fjyB_
not modelled
99.6
15
PDB header: hydrolaseChain: B: PDB Molecule: probable mutt1 protein;PDBTitle: crystal structure of a probable mutt1 protein from bifidobacterium2 adolescentis
47 c4hbzA_
not modelled
99.5
21
PDB header: hydrolase, isomeraseChain: A: PDB Molecule: putative phosphohistidine phosphatase, sixa;PDBTitle: the structure of putative phosphohistidine phosphatase sixa from2 nakamurella multipartitia.
48 c2zj6A_
not modelled
99.3
11
PDB header: hydrolaseChain: A: PDB Molecule: lipase;PDBTitle: crystal structure of d337a mutant of pseudomonas sp. mis38 lipase
49 c2qubG_
not modelled
99.3
13
PDB header: hydrolaseChain: G: PDB Molecule: extracellular lipase;PDBTitle: crystal structure of extracellular lipase lipa from serratia2 marcescens
50 c1k7qA_
not modelled
99.0
11
PDB header: hydrolaseChain: A: PDB Molecule: secreted protease c;PDBTitle: prtc from erwinia chrysanthemi: e189a mutant
51 c1satA_
not modelled
98.5
18
PDB header: hydrolase (serine protease)Chain: A: PDB Molecule: serratia protease;PDBTitle: crystal structure of the 50 kda metallo protease from s.2 marcescens
52 c3p4gD_
not modelled
98.5
11
PDB header: antifreeze proteinChain: D: PDB Molecule: antifreeze protein;PDBTitle: x-ray crystal structure of a hyperactive, ca2+-dependent, beta-helical2 antifreeze protein from an antarctic bacterium
53 d1kapp1
not modelled
98.1
17
Fold: Single-stranded right-handed beta-helixSuperfamily: beta-RollFamily: Serralysin-like metalloprotease, C-terminal domain
54 c1ygvA_
not modelled
98.1
15
PDB header: structural protein/contractile proteinChain: A: PDB Molecule: collagen i alpha 1;PDBTitle: the structure of collagen type i. single type i collagen2 molecule: rigid refinment
55 d1sata1
not modelled
98.0
15
Fold: Single-stranded right-handed beta-helixSuperfamily: beta-RollFamily: Serralysin-like metalloprotease, C-terminal domain
56 d1g9ka1
not modelled
97.9
15
Fold: Single-stranded right-handed beta-helixSuperfamily: beta-RollFamily: Serralysin-like metalloprotease, C-terminal domain
57 c1om8A_
not modelled
97.9
22
PDB header: hydrolaseChain: A: PDB Molecule: serralysin;PDBTitle: crystal structure of a cold adapted alkaline protease from pseudomonas2 tac ii 18, co-crystallyzed with 10 mm edta
58 d1k7ia1
not modelled
97.8
18
Fold: Single-stranded right-handed beta-helixSuperfamily: beta-RollFamily: Serralysin-like metalloprotease, C-terminal domain
59 c1jiwP_
not modelled
97.7
22
PDB header: hydrolase/hyrolase inhibitorChain: P: PDB Molecule: alkaline metalloproteinase;PDBTitle: crystal structure of the apr-aprin complex
60 c2ml3A_
not modelled
97.7
21
PDB header: isomeraseChain: A: PDB Molecule: poly(beta-d-mannuronate) c5 epimerase 6;PDBTitle: solution structure of alge6r3 subunit from the azotobacter vinelandii2 mannuronan c5-epimerase
61 c2ml2A_
not modelled
97.5
15
PDB header: isomeraseChain: A: PDB Molecule: poly(beta-d-mannuronate) c5 epimerase 6;PDBTitle: solution structure of alge6r2 subunit from the azotobacter vinelandii2 mannuronan c5-epimerase
62 c2agmA_
not modelled
97.4
24
PDB header: isomeraseChain: A: PDB Molecule: poly(beta-d-mannuronate) c5 epimerase 4;PDBTitle: solution structure of the r-module from alge4
63 c3hqvB_
not modelled
97.2
15
PDB header: structural protein, contractile proteinChain: B: PDB Molecule: collagen alpha-2(i) chain;PDBTitle: low resolution, molecular envelope structure of type i2 collagen in situ determined by fiber diffraction. single3 type i collagen molecule, rigid body refinement
64 c4q1qA_
not modelled
96.6
12
PDB header: cell adhesionChain: A: PDB Molecule: adhesin/invasin tiba autotransporter;PDBTitle: crystal structure of tibc-catalyzed hyper-glycosylated tiba55-3502 fragment
65 c5xfsA_
not modelled
96.4
53
PDB header: protein transportChain: A: PDB Molecule: pe family protein pe8;PDBTitle: crystal structure of pe8-ppe15 in complex with espg5 from m.2 tuberculosis
66 c2g38A_
not modelled
96.4
35
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: pe family protein;PDBTitle: a pe/ppe protein complex from mycobacterium tuberculosis
67 d2g38a1
not modelled
96.4
35
Fold: Ferritin-likeSuperfamily: PE/PPE dimer-likeFamily: PE
68 c1y0fB_
not modelled
96.3
16
PDB header: structural protein/contractile proteinChain: B: PDB Molecule: collagen i alpha 2;PDBTitle: the structure of collagen type i. single type i collagen2 molecule
69 c5cxlA_
not modelled
96.2
25
PDB header: toxinChain: A: PDB Molecule: bifunctional hemolysin/adenylate cyclase;PDBTitle: crystal structure of rtx domain block v of adenylate cyclase toxin2 from bordetella pertussis
70 c2glcA_
not modelled
91.3
24
PDB header: hydrolaseChain: A: PDB Molecule: histidine acid phosphatase;PDBTitle: structure of francisella tularensis histidine acid phosphatase bound2 to orthovanadate
71 c5cdhE_
not modelled
89.3
28
PDB header: hydrolase/hydrolase inhibitorChain: E: PDB Molecule: major acid phosphatase;PDBTitle: structure of legionella pneumophila histidine acid phosphatase2 complexed with l(+)-tartrate
72 c5juhA_
not modelled
81.3
11
PDB header: cell adhesionChain: A: PDB Molecule: antifreeze protein;PDBTitle: crystal structure of c-terminal domain (rv) of mpafp
73 c4jodA_
not modelled
80.8
25
PDB header: hydrolaseChain: A: PDB Molecule: lysophosphatidic acid phosphatase type 6;PDBTitle: crystal structure of human lysophosphatidic acid phosphatase type 62 complexed with tris
74 d1nd6a_
not modelled
76.6
26
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Histidine acid phosphatase
75 d1ihpa_
not modelled
75.0
16
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Histidine acid phosphatase
76 d1rpaa_
not modelled
72.7
21
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Histidine acid phosphatase
77 c4fdtB_
not modelled
71.7
18
PDB header: hydrolaseChain: B: PDB Molecule: putative multiple inositol polyphosphate histidinePDBTitle: crystal structure of a multiple inositol polyphosphate phosphatase
78 c4aruA_
not modelled
71.4
21
PDB header: hydrolaseChain: A: PDB Molecule: histidine acid phosphatase;PDBTitle: hafnia alvei phytase in complex with tartrate
79 d1qfxa_
not modelled
67.7
23
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Histidine acid phosphatase
80 c3zhcB_
not modelled
67.3
20
PDB header: hydrolaseChain: B: PDB Molecule: phytase;PDBTitle: structure of the phytase from citrobacter braakii at 2.3 angstrom2 resolution.
81 c4arvB_
not modelled
66.0
22
PDB header: hydrolaseChain: B: PDB Molecule: phytase;PDBTitle: yersinia kristensenii phytase apo form
82 c2pneA_
not modelled
60.7
36
PDB header: antifreeze proteinChain: A: PDB Molecule: 6.5 kda glycine-rich antifreeze protein;PDBTitle: crystal structure of the snow flea antifreeze protein
83 c3boiA_
not modelled
60.7
36
PDB header: antifreeze proteinChain: A: PDB Molecule: 6.5 kda glycine-rich antifreeze protein;PDBTitle: snow flea antifreeze protein racemate
84 c3boiB_
not modelled
60.7
36
PDB header: antifreeze proteinChain: B: PDB Molecule: 6.5 kda glycine-rich antifreeze protein;PDBTitle: snow flea antifreeze protein racemate
85 c3bogA_
not modelled
57.2
45
PDB header: antifreeze proteinChain: A: PDB Molecule: 6.5 kda glycine-rich antifreeze protein;PDBTitle: snow flea antifreeze protein quasi-racemate
86 c3bogB_
not modelled
57.2
45
PDB header: antifreeze proteinChain: B: PDB Molecule: 6.5 kda glycine-rich antifreeze protein;PDBTitle: snow flea antifreeze protein quasi-racemate
87 d1nt4a_
not modelled
55.3
20
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Histidine acid phosphatase
88 c2gfiB_
not modelled
55.1
23
PDB header: hydrolaseChain: B: PDB Molecule: phytase;PDBTitle: crystal structure of the phytase from d. castellii at 2.3 a
89 d1qwoa_
not modelled
52.8
20
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Histidine acid phosphatase
90 c2wniC_
not modelled
47.9
24
PDB header: hydrolaseChain: C: PDB Molecule: 3-phytase;PDBTitle: crystal structure analysis of klebsiella sp asr1 phytase
91 d1dkla_
not modelled
42.9
26
Fold: Phosphoglycerate mutase-likeSuperfamily: Phosphoglycerate mutase-likeFamily: Histidine acid phosphatase
92 c3m4xA_
not modelled
42.9
43
PDB header: transferaseChain: A: PDB Molecule: nol1/nop2/sun family protein;PDBTitle: structure of a ribosomal methyltransferase
93 c3gzrA_
not modelled
33.7
28
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein with a ntf2-like fold;PDBTitle: crystal structure of an uncharacterized protein with a cystatin-like2 fold (cc_2572) from caulobacter vibrioides at 1.40 a resolution
94 c2frxD_
not modelled
32.2
32
PDB header: transferaseChain: D: PDB Molecule: hypothetical protein yebu;PDBTitle: crystal structure of yebu, a m5c rna methyltransferase from e.coli
95 c2eqoA_
not modelled
24.2
24
PDB header: transcriptionChain: A: PDB Molecule: tnf receptor-associated factor 3-interactingPDBTitle: solution structure of the stn_traf3ip1_nd domain of2 interleukin 13 receptor alpha 1-binding protein-1 [homo3 sapiens]
96 c5fmtB_
not modelled
24.1
33
PDB header: protein transportChain: B: PDB Molecule: flagellar associated protein;PDBTitle: crift54 ch-domain
97 d2qn6a2
not modelled
19.1
22
Fold: Elongation factor/aminomethyltransferase common domainSuperfamily: EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domainFamily: EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain
98 c2l3aA_
not modelled
18.5
40
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of homodimer protein sp_0782 (7-79) from2 streptococcus pneumoniae northeast structural genomics consortium3 target spr104 .
99 c2ltdA_
not modelled
18.2
35
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ydbc;PDBTitle: solution nmr structure of apo ydbc from lactococcus lactis, northeast2 structural genomics consortium (nesg) target kr150