| 1 |
|
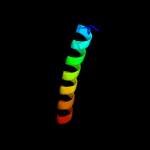
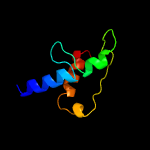
PDB 4wpy chain A
Region: 72 - 105
Aligned: 34
Modelled: 34
Confidence: 28.3%
Identity: 18%
PDB header:de novo protein
Chain: A: PDB Molecule:protein dl-rv1738;
PDBTitle: racemic crystal structure of rv1738 from mycobacterium tuberculosis2 (form-ii)
Phyre2
| 2 |
|
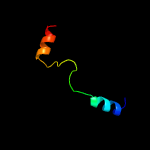
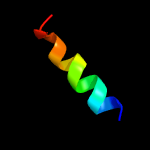
PDB 1gr0 chain A domain 1
Region: 73 - 88
Aligned: 16
Modelled: 16
Confidence: 27.0%
Identity: 44%
Fold: NAD(P)-binding Rossmann-fold domains
Superfamily: NAD(P)-binding Rossmann-fold domains
Family: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
Phyre2
| 3 |
|
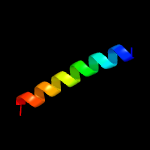
PDB 2fgg chain A domain 1
Region: 73 - 105
Aligned: 33
Modelled: 33
Confidence: 26.3%
Identity: 15%
Fold: dsRBD-like
Superfamily: Rv2632c-like
Family: Rv2632c-like
Phyre2
| 4 |
|
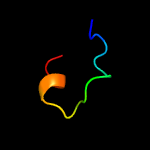
PDB 5ydc chain A
Region: 172 - 204
Aligned: 31
Modelled: 33
Confidence: 23.8%
Identity: 19%
PDB header:transcription regulator
Chain: A: PDB Molecule:uncharacterized hth-type transcriptional regulator rv1828;
PDBTitle: crystal structure of mercury soaked c-terminal domain of rv1828 from2 mycobacterium tuberculosis
Phyre2
| 5 |
|
PDB 2axt chain M domain 1
Region: 106 - 130
Aligned: 25
Modelled: 25
Confidence: 17.4%
Identity: 8%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction center protein M, PsbM
Family: PsbM-like
Phyre2
| 6 |
|
PDB 3a0h chain M
Region: 106 - 130
Aligned: 25
Modelled: 25
Confidence: 14.3%
Identity: 8%
PDB header:electron transport
Chain: M: PDB Molecule:photosystem ii reaction center protein m;
PDBTitle: crystal structure of i-substituted photosystem ii complex
Phyre2
| 7 |
|
PDB 1q90 chain B
Region: 26 - 130
Aligned: 86
Modelled: 92
Confidence: 11.8%
Identity: 20%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 8 |
|
PDB 2l35 chain B
Region: 8 - 35
Aligned: 28
Modelled: 28
Confidence: 11.2%
Identity: 21%
PDB header:protein binding
Chain: B: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12-nkg2c transmembrane heterotrimer
Phyre2
| 9 |
|
PDB 2l34 chain B
Region: 8 - 35
Aligned: 28
Modelled: 28
Confidence: 11.0%
Identity: 21%
PDB header:protein binding
Chain: B: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12 transmembrane homodimer
Phyre2
| 10 |
|
PDB 4wol chain A
Region: 8 - 35
Aligned: 28
Modelled: 28
Confidence: 11.0%
Identity: 21%
PDB header:signaling protein
Chain: A: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: crystal structure of the dap12 transmembrane domain in lipidic cubic2 phase
Phyre2
| 11 |
|
PDB 2l34 chain A
Region: 8 - 35
Aligned: 28
Modelled: 28
Confidence: 11.0%
Identity: 21%
PDB header:protein binding
Chain: A: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12 transmembrane homodimer
Phyre2
| 12 |
|
PDB 4h1w chain B
Region: 118 - 133
Aligned: 16
Modelled: 16
Confidence: 8.4%
Identity: 25%
PDB header:hydrolase/hydrolase regulator
Chain: B: PDB Molecule:sarcolipin;
PDBTitle: e1 structure of the (sr) ca2+-atpase in complex with sarcolipin
Phyre2
| 13 |
|
PDB 3w5a chain C
Region: 118 - 133
Aligned: 16
Modelled: 16
Confidence: 8.4%
Identity: 25%
PDB header:metal transport/membrane protein
Chain: C: PDB Molecule:sarcolipin;
PDBTitle: crystal structure of the calcium pump and sarcolipin from rabbit fast2 twitch skeletal muscle in the e1.mg2+ state
Phyre2
| 14 |
|
PDB 1vp5 chain A
Region: 161 - 194
Aligned: 34
Modelled: 34
Confidence: 8.2%
Identity: 18%
Fold: TIM beta/alpha-barrel
Superfamily: NAD(P)-linked oxidoreductase
Family: Aldo-keto reductases (NADP)
Phyre2
| 15 |
|
PDB 1k6n chain H
Region: 22 - 45
Aligned: 24
Modelled: 24
Confidence: 7.5%
Identity: 8%
PDB header:photosynthesis
Chain: H: PDB Molecule:photosynthetic reaction center h subunit;
PDBTitle: e(l212)a,d(l213)a double mutant structure of photosynthetic reaction2 center from rhodobacter sphaeroides
Phyre2
| 16 |
|
PDB 4v1a chain Q
Region: 165 - 182
Aligned: 18
Modelled: 18
Confidence: 6.9%
Identity: 33%
PDB header:ribosome
Chain: Q: PDB Molecule:
PDBTitle: structure of the large subunit of the mammalian mitoribosome, part 22 of 2
Phyre2
| 17 |
|
PDB 1eys chain H
Region: 22 - 45
Aligned: 24
Modelled: 24
Confidence: 5.9%
Identity: 8%
PDB header:electron transport
Chain: H: PDB Molecule:photosynthetic reaction center;
PDBTitle: crystal structure of photosynthetic reaction center from a2 thermophilic bacterium, thermochromatium tepidum
Phyre2
| 18 |
|
PDB 1nbw chain A domain 3
Region: 133 - 201
Aligned: 61
Modelled: 69
Confidence: 5.8%
Identity: 20%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: ATPase domain of dehydratase reactivase alpha subunit
Phyre2
| 19 |
|
PDB 2i5n chain H
Region: 22 - 36
Aligned: 15
Modelled: 15
Confidence: 5.7%
Identity: 33%
PDB header:photosynthesis
Chain: H: PDB Molecule:reaction center protein h chain;
PDBTitle: 1.96 a x-ray structure of photosynthetic reaction center from2 rhodopseudomonas viridis:crystals grown by microfluidic technique
Phyre2
| 20 |
|
PDB 1yyb chain A
Region: 158 - 176
Aligned: 19
Modelled: 19
Confidence: 5.5%
Identity: 37%
PDB header:apoptosis
Chain: A: PDB Molecule:programmed cell death protein 5;
PDBTitle: solution structure of 1-26 fragment of human programmed2 cell death 5 protein
Phyre2
| 21 |
|