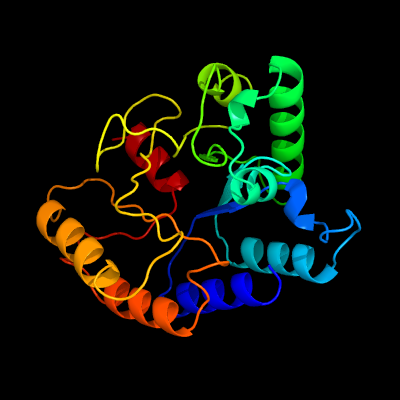
| 1 | c3qbuD_
|
|
 |
100.0 |
14 |
PDB header:hydrolase
Chain: D: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of putative peptidoglycan deactelyase (hp0310) from2 helicobacter pylori
|
|
|
|
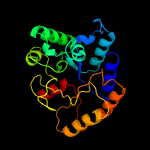
| 2 | d1z7aa1
|
|
 |
100.0 |
13 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:PA1517-like |
|
|
|
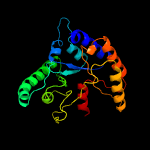
| 3 | c3rxzA_
|
|
 |
100.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:polysaccharide deacetylase;
PDBTitle: crystal structure of putative polysaccharide deacetylase from2 mycobacterium smegmatis
|
|
|
|
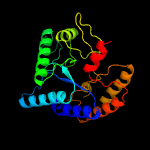
| 4 | c3s6oD_
|
|
 |
100.0 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:polysaccharide deacetylase family protein;
PDBTitle: crystal structure of a polysaccharide deacetylase family protein from2 burkholderia pseudomallei
|
|
|
|
| 5 | c2c1iA_
|
|
 |
100.0 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidoglycan glcnac deacetylase;
PDBTitle: structure of streptococcus pneumoniae peptidoglycan deacetylase2 (sppgda) d 275 n mutant.
|
|
|
|
| 6 | c4nz3A_
|
|
 |
100.0 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:deacetylase da1;
PDBTitle: structure of vibrio cholerae chitin de-n-acetylase in complex with2 di(n-acetyl-d-glucosamine) (cbs) in p 21 21 21
|
|
|
|
| 7 | c2w3zA_
|
|
 |
100.0 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative deacetylase;
PDBTitle: structure of a streptococcus mutans ce4 esterase
|
|
|
|
| 8 | c1w17A_
|
|
 |
100.0 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable polysaccharide deacetylase pdaa;
PDBTitle: structure of bacillus subtilis pdaa, a family 4 carbohydrate esterase.
|
|
|
|
| 9 | c5jp6A_
|
|
 |
100.0 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative polysaccharide deacetylase;
PDBTitle: bdellovibrio bacteriovorus peptidoglycan deacetylase bd3279
|
|
|
|
| 10 | c4m1bA_
|
|
 |
100.0 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:polysaccharide deacetylase;
PDBTitle: structural determination of ba0150, a polysaccharide deacetylase from2 bacillus anthracis
|
|
|
|
| 11 | d1ny1a_
|
|
 |
100.0 |
12 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:NodB-like polysaccharide deacetylase |
|
|
|
| 12 | d2cc0a1
|
|
 |
100.0 |
12 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:NodB-like polysaccharide deacetylase |
|
|
|
| 13 | d2iw0a1
|
|
 |
100.0 |
10 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:NodB-like polysaccharide deacetylase |
|
|
|
| 14 | c5lgcA_
|
|
 |
100.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:arce4a;
PDBTitle: t48 deacetylase with substrate
|
|
|
|
| 15 | c4l1gB_
|
|
 |
100.0 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:peptidoglycan n-acetylglucosamine deacetylase;
PDBTitle: crystal structure of the bc1960 peptidoglycan n-acetylglucosamine2 deacetylase from bacillus cereus
|
|
|
|
| 16 | c5ncdA_
|
|
 |
100.0 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidoglycan n-acetylglucosamine deacetylase;
PDBTitle: crystal structure of the polysaccharide deacetylase bc1974 from2 bacillus cereus in complex with (2s)-2-amino-5-3 (diaminomethylideneamino)-n-hydroxypentanamide
|
|
|
|
| 17 | d2j13a1
|
|
 |
100.0 |
13 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:NodB-like polysaccharide deacetylase |
|
|
|
| 18 | c2iw0A_
|
|
 |
99.9 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:chitin deacetylase;
PDBTitle: structure of the chitin deacetylase from the fungal2 pathogen colletotrichum lindemuthianum
|
|
|
|
| 19 | d2c71a1
|
|
 |
99.9 |
14 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:NodB-like polysaccharide deacetylase |
|
|
|
| 20 | d2c1ia1
|
|
 |
99.9 |
13 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:NodB-like polysaccharide deacetylase |
|
|
|
| 21 | c2y8uA_ |
|
not modelled |
99.9 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:chitin deacetylase;
PDBTitle: a. nidulans chitin deacetylase
|
|
|
| 22 | c2vyoA_ |
|
not modelled |
99.9 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:polysaccharide deacetylase domain-containing protein
PDBTitle: chitin deacetylase family member from encephalitozoon cuniculi
|
|
|
| 23 | c5z34A_ |
|
not modelled |
99.9 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:chitin deacetylase;
PDBTitle: the structure of a chitin deacetylase from bombyx mori provide the2 first insight into insect chitin deacetylation mechanism
|
|
|
| 24 | d2nlya1 |
|
not modelled |
99.9 |
14 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:Divergent polysaccharide deacetylase |
|
|
| 25 | c5znsA_ |
|
not modelled |
99.9 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:chitin deacetylase;
PDBTitle: insect chitin deacetylase
|
|
|
| 26 | c4wcjA_ |
|
not modelled |
99.8 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:polysaccharide deacetylase;
PDBTitle: structure of icab from ammonifex degensii
|
|
|
| 27 | c6dq3B_ |
|
not modelled |
99.8 |
9 |
PDB header:hydrolase
Chain: B: PDB Molecule:polysaccharide deacetylase;
PDBTitle: streptococcus pyogenes deacetylase pdi in complex with acetate
|
|
|
| 28 | c4f9dA_ |
|
not modelled |
99.7 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:poly-beta-1,6-n-acetyl-d-glucosamine n-deacetylase;
PDBTitle: structure of escherichia coli pgab 42-655 in complex with nickel
|
|
|
| 29 | c5bu6B_ |
|
not modelled |
99.7 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:bpsb (pgab), poly-beta-1,6-n-acetyl-d-glucosamine n-
PDBTitle: structure of bpsb deaceylase domain from bordetella bronchiseptica
|
|
|
| 30 | c4u10B_ |
|
not modelled |
99.6 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:poly-beta-1,6-n-acetyl-d-glucosamine n-deacetylase;
PDBTitle: probing the structure and mechanism of de-n-acetylase from2 aggregatibacter actinomycetemcomitans
|
|
|
| 31 | c6go1A_ |
|
not modelled |
99.4 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:polysaccharide deacetylase-like protein;
PDBTitle: crystal structure of a bacillus anthracis peptidoglycan deacetylase
|
|
|
| 32 | c4hd5A_ |
|
not modelled |
99.1 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:polysaccharide deacetylase;
PDBTitle: crystal structure of bc0361, a polysaccharide deacetylase from2 bacillus cereus
|
|
|
| 33 | d1k1xa3 |
|
not modelled |
99.0 |
12 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:4-alpha-glucanotransferase, N-terminal domain |
|
|
| 34 | c1k1yA_ |
|
not modelled |
98.8 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:4-alpha-glucanotransferase;
PDBTitle: crystal structure of thermococcus litoralis 4-alpha-glucanotransferase2 complexed with acarbose
|
|
|
| 35 | d2b5dx2 |
|
not modelled |
98.5 |
14 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:AmyC N-terminal domain-like |
|
|
| 36 | c2b5dX_ |
|
not modelled |
98.4 |
15 |
PDB header:hydrolase
Chain: X: PDB Molecule:alpha-amylase;
PDBTitle: crystal structure of the novel alpha-amylase amyc from thermotoga2 maritima
|
|
|
| 37 | c2qv5A_ |
|
not modelled |
98.3 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein atu2773;
PDBTitle: crystal structure of uncharacterized protein atu2773 from2 agrobacterium tumefaciens c58
|
|
|
| 38 | c5wu7A_ |
|
not modelled |
97.8 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of gh57-type branching enzyme from hyperthermophilic2 archaeon pyrococcus horikoshii
|
|
|
| 39 | c3n92A_ |
|
not modelled |
97.5 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:alpha-amylase, gh57 family;
PDBTitle: crystal structure of tk1436, a gh57 branching enzyme from2 hyperthermophilic archaeon thermococcus kodakaraensis, in complex3 with glucose
|
|
|
| 40 | c1ufaA_ |
|
not modelled |
97.1 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:tt1467 protein;
PDBTitle: crystal structure of tt1467 from thermus thermophilus hb8
|
|
|
| 41 | d1ufaa2 |
|
not modelled |
96.9 |
15 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:AmyC N-terminal domain-like |
|
|
| 42 | c2wyhA_ |
|
not modelled |
96.4 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-mannosidase;
PDBTitle: structure of the streptococcus pyogenes family gh38 alpha-2 mannosidase
|
|
|
| 43 | c3hftA_ |
|
not modelled |
96.3 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:wbms, polysaccharide deacetylase involved in o-antigen
PDBTitle: crystal structure of a putative polysaccharide deacetylase involved in2 o-antigen biosynthesis (wbms, bb0128) from bordetella bronchiseptica3 at 1.90 a resolution
|
|
|
| 44 | c2ow7A_ |
|
not modelled |
93.2 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-mannosidase 2;
PDBTitle: golgi alpha-mannosidase ii complex with (1r,6s,7r,8s)-1-2 thioniabicyclo[4.3.0]nonan-7,8-diol chloride
|
|
|
| 45 | c1o7dA_ |
|
not modelled |
93.1 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:lysosomal alpha-mannosidase;
PDBTitle: the structure of the bovine lysosomal a-mannosidase suggests a novel2 mechanism for low ph activation
|
|
|
| 46 | c2hk1D_ |
|
not modelled |
92.7 |
7 |
PDB header:isomerase
Chain: D: PDB Molecule:d-psicose 3-epimerase;
PDBTitle: crystal structure of d-psicose 3-epimerase (dpease) in the presence of2 d-fructose
|
|
|
| 47 | c1htyA_ |
|
not modelled |
92.5 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-mannosidase ii;
PDBTitle: golgi alpha-mannosidase ii
|
|
|
| 48 | d2b7oa1 |
|
not modelled |
91.8 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class-II DAHP synthetase |
|
|
| 49 | c5hudA_ |
|
not modelled |
91.2 |
17 |
PDB header:transferase/isomerase
Chain: A: PDB Molecule:3-deoxy-d-arabino-heptulosonate 7-phosphate (dahp)
PDBTitle: non-covalent complex of and dahp synthase and chorismate mutase from2 corynebacterium glutamicum with bound transition state analog
|
|
|
| 50 | c6bmcA_ |
|
not modelled |
91.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:phospho-2-dehydro-3-deoxyheptonate aldolase;
PDBTitle: the structure of a dimeric type ii dah7ps associated with pyocyanin2 biosynthesis in pseudomonas aeruginosa
|
|
|
| 51 | c5uxmA_ |
|
not modelled |
90.9 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:phospho-2-dehydro-3-deoxyheptonate aldolase;
PDBTitle: type ii dah7ps from pseudomonas aeruginosa with trp bound
|
|
|
| 52 | d3bvua3 |
|
not modelled |
90.3 |
16 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:alpha-mannosidase |
|
|
| 53 | c2o7vA_ |
|
not modelled |
89.2 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:cxe carboxylesterase;
PDBTitle: carboxylesterase aecxe1 from actinidia eriantha covalently inhibited2 by paraoxon
|
|
|
| 54 | d1i60a_ |
|
not modelled |
88.1 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
|
|
| 55 | c5zfsA_ |
|
not modelled |
85.7 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:d-allulose-3-epimerase;
PDBTitle: crystal structure of arthrobacter globiformis m30 sugar epimerase2 which can produce d-allulose from d-fructose
|
|
|
| 56 | c3ktcB_ |
|
not modelled |
83.7 |
8 |
PDB header:isomerase
Chain: B: PDB Molecule:xylose isomerase;
PDBTitle: crystal structure of putative sugar isomerase (yp_050048.1) from2 erwinia carotovora atroseptica scri1043 at 1.54 a resolution
|
|
|
| 57 | c3kwsB_ |
|
not modelled |
82.3 |
9 |
PDB header:isomerase
Chain: B: PDB Molecule:putative sugar isomerase;
PDBTitle: crystal structure of putative sugar isomerase (yp_001305149.1) from2 parabacteroides distasonis atcc 8503 at 1.68 a resolution
|
|
|
| 58 | c6b9pA_ |
|
not modelled |
81.4 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-mannosidase from canavalia ensiformis (jack bean);
PDBTitle: structure of gh 38 jack bean alpha-mannosidase in complex with a 36-2 valent iminosugar cluster inhibitor
|
|
|
| 59 | c5oljA_ |
|
not modelled |
79.3 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:dipeptidyl peptidase iv;
PDBTitle: crystal structure of porphyromonas gingivalis dipeptidyl peptidase 4
|
|
|
| 60 | d2g0wa1 |
|
not modelled |
77.6 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
|
|
| 61 | d1muwa_ |
|
not modelled |
77.5 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
|
|
| 62 | d1u4na_ |
|
not modelled |
77.4 |
7 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Carboxylesterase |
|
|
| 63 | c3obeB_ |
|
not modelled |
76.5 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:sugar phosphate isomerase/epimerase;
PDBTitle: crystal structure of a sugar phosphate isomerase/epimerase (bdi_3400)2 from parabacteroides distasonis atcc 8503 at 1.70 a resolution
|
|
|
| 64 | c4ob7A_ |
|
not modelled |
75.9 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha/beta hydrolase fold-3 domain protein;
PDBTitle: crystal structure of esterase rppe mutant w187h
|
|
|
| 65 | c4v2iB_ |
|
not modelled |
75.7 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:esterase/lipase;
PDBTitle: biochemical characterization and structural analysis of a2 new cold-active and salt tolerant esterase from the marine3 bacterium thalassospira sp
|
|
|
| 66 | c3d7rB_ |
|
not modelled |
74.6 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:esterase;
PDBTitle: crystal structure of a putative esterase from staphylococcus aureus
|
|
|
| 67 | c3cqkB_ |
|
not modelled |
74.4 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:l-ribulose-5-phosphate 3-epimerase ulae;
PDBTitle: crystal structure of l-xylulose-5-phosphate 3-epimerase ulae (form b)2 complex with zn2+ and sulfate
|
|
|
| 68 | d1bxca_ |
|
not modelled |
73.8 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
|
|
| 69 | d1jjia_ |
|
not modelled |
72.6 |
9 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Carboxylesterase |
|
|
| 70 | c2ou4C_ |
|
not modelled |
72.0 |
10 |
PDB header:isomerase
Chain: C: PDB Molecule:d-tagatose 3-epimerase;
PDBTitle: crystal structure of d-tagatose 3-epimerase from2 pseudomonas cichorii
|
|
|
| 71 | c5jd5A_ |
|
not modelled |
71.7 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:mgs-mile3;
PDBTitle: crystal structure of mgs-mile3, an alpha/beta hydrolase enzyme from2 the metagenome of pyrene-phenanthrene enrichment culture with3 sediment sample of milazzo harbor, italy
|
|
|
| 72 | c3vylB_ |
|
not modelled |
71.1 |
10 |
PDB header:isomerase
Chain: B: PDB Molecule:l-ribulose 3-epimerase;
PDBTitle: structure of l-ribulose 3-epimerase
|
|
|
| 73 | c2g5tA_ |
|
not modelled |
70.7 |
8 |
PDB header:hydrolase
Chain: A: PDB Molecule:dipeptidyl peptidase 4;
PDBTitle: crystal structure of human dipeptidyl peptidase iv (dppiv) complexed2 with cyanopyrrolidine (c5-pro-pro) inhibitor 21ag
|
|
|
| 74 | c4krxB_ |
|
not modelled |
70.6 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:acetyl esterase;
PDBTitle: structure of aes from e. coli
|
|
|
| 75 | c2f46A_ |
|
not modelled |
70.6 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a putative phosphatase (nma1982) from neisseria2 meningitidis z2491 at 1.41 a resolution
|
|
|
| 76 | c3aikB_ |
|
not modelled |
69.4 |
7 |
PDB header:hydrolase
Chain: B: PDB Molecule:303aa long hypothetical esterase;
PDBTitle: crystal structure of a hsl-like carboxylesterase from sulfolobus2 tokodaii
|
|
|
| 77 | c4xvcG_ |
|
not modelled |
68.9 |
20 |
PDB header:hydrolase
Chain: G: PDB Molecule:esterase e40;
PDBTitle: crystal structure of an esterase from the bacterial hormone-sensitive2 lipase (hsl) family
|
|
|
| 78 | c3ed1E_ |
|
not modelled |
67.3 |
9 |
PDB header:hydrolase receptor
Chain: E: PDB Molecule:gibberellin receptor gid1;
PDBTitle: crystal structure of rice gid1 complexed with ga3
|
|
|
| 79 | c3fakA_ |
|
not modelled |
66.7 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:esterase/lipase;
PDBTitle: structural and functional analysis of a hormone-sensitive2 lipase like este5 from a metagenome library
|
|
|
| 80 | c5l8sD_ |
|
not modelled |
65.9 |
13 |
PDB header:hydrolase
Chain: D: PDB Molecule:amino acyl peptidase;
PDBTitle: the crystal structure of a cold-adapted acylaminoacyl peptidase2 reveals a novel quaternary architecture based on the arm-exchange3 mechanism
|
|
|
| 81 | c6aaeA_ |
|
not modelled |
65.6 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:esterase;
PDBTitle: crystal structure of chloramphenicol-metabolizaing enzyme estdl136
|
|
|
| 82 | c2wirB_ |
|
not modelled |
64.6 |
5 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha/beta hydrolase fold-3 domain protein;
PDBTitle: hyperthermophilic esterase from the archeon pyrobaculum2 calidifontis
|
|
|
| 83 | c5jd4D_ |
|
not modelled |
63.9 |
5 |
PDB header:hydrolase
Chain: D: PDB Molecule:lae6;
PDBTitle: crystal structure of lae6 ser161ala mutant, an alpha/beta hydrolase2 enzyme from the metagenome of lake arreo, spain
|
|
|
| 84 | c6gl2A_ |
|
not modelled |
63.9 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:endoglucanase, family gh5;
PDBTitle: structure of zgengagh5_4 wild type at 1.2 angstrom resolution
|
|
|
| 85 | c4q05A_ |
|
not modelled |
63.7 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:esterase e25;
PDBTitle: crystal structure of an esterase e25
|
|
|
| 86 | c5miiD_ |
|
not modelled |
63.1 |
9 |
PDB header:hydrolase
Chain: D: PDB Molecule:carboxyl esterase 2;
PDBTitle: crystal structure of carboxyl esterase 2 (tmelest2) from mycorrhizal2 fungus tuber melanosporum
|
|
|
| 87 | c4q3oB_ |
|
not modelled |
62.9 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:mgs-mt1;
PDBTitle: crystal structure of mgs-mt1, an alpha/beta hydrolase enzyme from a2 lake matapan deep-sea metagenome library
|
|
|
| 88 | c2ecfA_ |
|
not modelled |
62.7 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:dipeptidyl peptidase iv;
PDBTitle: crystal structure of dipeptidyl aminopeptidase iv from2 stenotrophomonas maltophilia
|
|
|
| 89 | c1vjqB_ |
|
not modelled |
62.1 |
22 |
PDB header:structural genomics, de novo protein
Chain: B: PDB Molecule:designed protein;
PDBTitle: designed protein based on backbone conformation of2 procarboxypeptidase-a (1aye) with sidechains chosen for maximal3 predicted stability.
|
|
|
| 90 | c3wj2A_ |
|
not modelled |
62.0 |
7 |
PDB header:hydrolase
Chain: A: PDB Molecule:carboxylesterase;
PDBTitle: crystal structure of estfa (fe-lacking apo form)
|
|
|
| 91 | c4wy8A_ |
|
not modelled |
61.9 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:esterase;
PDBTitle: structural analysis of two fungal esterases from rhizomucor miehei2 explaining their substrate specificity
|
|
|
| 92 | c2qtbB_ |
|
not modelled |
60.5 |
8 |
PDB header:hydrolase
Chain: B: PDB Molecule:dipeptidyl peptidase 4;
PDBTitle: human dipeptidyl peptidase iv/cd26 in complex with a 4-aryl2 cyclohexylalanine inhibitor
|
|
|
| 93 | c3ju2A_ |
|
not modelled |
60.3 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein smc04130;
PDBTitle: crystal structure of protein smc04130 from sinorhizobium meliloti 1021
|
|
|
| 94 | c3ga7A_ |
|
not modelled |
59.9 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:acetyl esterase;
PDBTitle: 1.55 angstrom crystal structure of an acetyl esterase from salmonella2 typhimurium
|
|
|
| 95 | c3azqA_ |
|
not modelled |
59.0 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:aminopeptidase;
PDBTitle: crystal structure of puromycin hydrolase s511a mutant complexed with2 pgg
|
|
|
| 96 | d1lzla_ |
|
not modelled |
58.9 |
9 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Carboxylesterase |
|
|
| 97 | c4wy5A_ |
|
not modelled |
56.7 |
7 |
PDB header:hydrolase
Chain: A: PDB Molecule:esterase;
PDBTitle: structural analysis of two fungal esterases from rhizomucor miehei2 explaining their substrate specificity
|
|
|
| 98 | c5jm0A_ |
|
not modelled |
56.5 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-mannosidase,alpha-mannosidase,alpha-mannosidase;
PDBTitle: structure of the s. cerevisiae alpha-mannosidase 1
|
|
|
| 99 | c1xfdD_ |
|
not modelled |
55.9 |
6 |
PDB header:membrane protein
Chain: D: PDB Molecule:dipeptidyl aminopeptidase-like protein 6;
PDBTitle: structure of a human a-type potassium channel accelerating factor2 dppx, a member of the dipeptidyl aminopeptidase family
|
|
|
| 100 | c4ypvA_ |
|
not modelled |
54.5 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:est8;
PDBTitle: high-resolution structure of a metagenome-derived esterase est8
|
|
|
| 101 | c4zi5A_ |
|
not modelled |
53.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:p91;
PDBTitle: crystal structure of dienelactone hydrolase-like promiscuous2 phospotriesterase p91 from metagenomic libraries
|
|
|
| 102 | c3vniC_ |
|
not modelled |
51.3 |
8 |
PDB header:isomerase
Chain: C: PDB Molecule:xylose isomerase domain protein tim barrel;
PDBTitle: crystal structures of d-psicose 3-epimerase from clostridium2 cellulolyticum h10 and its complex with ketohexose sugars
|
|
|
| 103 | d1rxda_ |
|
not modelled |
51.2 |
16 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Dual specificity phosphatase-like |
|
|
| 104 | c3dnmA_ |
|
not modelled |
51.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:esterase/lipase;
PDBTitle: crystal structure hormone-sensitive lipase from a2 metagenome library
|
|
|
| 105 | c5l2pD_ |
|
not modelled |
50.5 |
17 |
PDB header:hydrolase
Chain: D: PDB Molecule:arylesterase;
PDBTitle: structure of arylesterase
|
|
|
| 106 | d2q02a1 |
|
not modelled |
46.8 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:IolI-like |
|
|
| 107 | d2i5ia1 |
|
not modelled |
46.4 |
15 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:YdjC-like |
|
|
| 108 | c4ovxA_ |
|
not modelled |
45.1 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:xylose isomerase domain protein tim barrel;
PDBTitle: crystal structure of xylose isomerase domain protein from planctomyces2 limnophilus dsm 3776
|
|
|
| 109 | c5jrlC_ |
|
not modelled |
44.8 |
10 |
PDB header:hydrolase
Chain: C: PDB Molecule:dipeptidyl aminopeptidases/acylaminoacyl-peptidases-like
PDBTitle: crystal structure of the sphingopyxin i lasso peptide isopeptidase2 spi-isop (native)
|
|
|
| 110 | c2k6xA_ |
|
not modelled |
44.4 |
12 |
PDB header:transcription
Chain: A: PDB Molecule:rna polymerase sigma factor rpod;
PDBTitle: autoregulation of a group 1 bacterial sigma factor involves2 the formation of a region 1.1- induced compacted structure
|
|
|
| 111 | c5yznA_ |
|
not modelled |
43.5 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:acyl-peptide hydrolase, putative;
PDBTitle: crystal structure of s9 peptidase (active form) from deinococcus2 radiodurans r1
|
|
|
| 112 | c3gk0H_ |
|
not modelled |
42.7 |
27 |
PDB header:transferase
Chain: H: PDB Molecule:pyridoxine 5'-phosphate synthase;
PDBTitle: crystal structure of pyridoxal phosphate biosynthetic protein from2 burkholderia pseudomallei
|
|
|
| 113 | d2pbdp1 |
|
not modelled |
40.9 |
0 |
Fold:Profilin-like
Superfamily:Profilin (actin-binding protein)
Family:Profilin (actin-binding protein) |
|
|
| 114 | c3cnyA_ |
|
not modelled |
40.2 |
13 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:inositol catabolism protein iole;
PDBTitle: crystal structure of a putative inositol catabolism protein iole2 (iole, lp_3607) from lactobacillus plantarum wcfs1 at 1.85 a3 resolution
|
|
|
| 115 | c1z68A_ |
|
not modelled |
39.8 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:fibroblast activation protein, alpha subunit;
PDBTitle: crystal structure of human fibroblast activation protein alpha
|
|
|
| 116 | d1pnea_ |
|
not modelled |
39.4 |
0 |
Fold:Profilin-like
Superfamily:Profilin (actin-binding protein)
Family:Profilin (actin-binding protein) |
|
|
| 117 | c2eepA_ |
|
not modelled |
39.4 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:dipeptidyl aminopeptidase iv, putative;
PDBTitle: prolyl tripeptidyl aminopeptidase complexed with an inhibitor
|
|
|
| 118 | c2zshA_ |
|
not modelled |
37.9 |
13 |
PDB header:hormone receptor
Chain: A: PDB Molecule:probable gibberellin receptor gid1l1;
PDBTitle: structural basis of gibberellin(ga3)-induced della2 recognition by the gibberellin receptor
|
|
|
| 119 | c5hc4A_ |
|
not modelled |
37.2 |
7 |
PDB header:hydrolase
Chain: A: PDB Molecule:lipolytic enzyme;
PDBTitle: structure of esterase est22
|
|
|
| 120 | c4wjlB_ |
|
not modelled |
36.8 |
8 |
PDB header:membrane protein
Chain: B: PDB Molecule:inactive dipeptidyl peptidase 10;
PDBTitle: structure of human dipeptidyl peptidase 10 (dppy): a modulator of2 neuronal kv4 channels
|
|
|