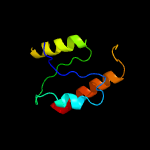
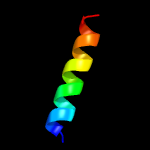
1 c3f5dA_
39.9
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein ydea;PDBTitle: crystal structure of a protein of unknown function from bacillus2 subtilis
2 c3c4rC_
37.8
28
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein encoded by2 cryptic prophage
3 d2bosa_
35.7
52
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits
4 d2fb5a1
34.8
27
Fold: YojJ-likeSuperfamily: YojJ-likeFamily: YojJ-like
5 d2auna2
31.3
20
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: LD-carboxypeptidase A N-terminal domain-like
6 c3n7tA_
28.2
10
PDB header: protein bindingChain: A: PDB Molecule: macrophage binding protein;PDBTitle: crystal structure of a macrophage binding protein from coccidioides2 immitis
7 c4rv7C_
26.2
27
PDB header: transferaseChain: C: PDB Molecule: diadenylate cyclase;PDBTitle: characterization of an essential diadenylate cyclase
8 c4lruA_
23.1
12
PDB header: lyaseChain: A: PDB Molecule: glyoxalase iii (glutathione-independent);PDBTitle: crystal structure of glyoxalase iii (orf 19.251) from candida albicans
9 c1trlB_
22.8
35
PDB header: hydrolase (metalloprotease)Chain: B: PDB Molecule: thermolysin fragment 255 - 316;PDBTitle: nmr solution structure of the c-terminal fragment 255-3162 of thermolysin: a dimer formed by subunits having the3 native structure
10 c3kklA_
22.2
14
PDB header: hydrolaseChain: A: PDB Molecule: probable chaperone protein hsp33;PDBTitle: crystal structure of functionally unknown hsp33 from2 saccharomyces cerevisiae
11 d1r4pb_
21.4
48
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits
12 d1gq2a2
20.5
19
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Malic enzyme N-domain
13 c6gyyB_
18.9
19
PDB header: transferaseChain: B: PDB Molecule: diadenylate cyclase;PDBTitle: crystal structure of daca from staphylococcus aureus, n166c/t172c2 double mutant
14 c3gdfA_
18.1
22
PDB header: oxidoreductaseChain: A: PDB Molecule: probable nadp-dependent mannitol dehydrogenase;PDBTitle: crystal structure of the nadp-dependent mannitol dehydrogenase from2 cladosporium herbarum.
15 d1d1da1
13.2
43
Fold: Acyl carrier protein-likeSuperfamily: Retrovirus capsid dimerization domain-likeFamily: Retrovirus capsid protein C-terminal domain
16 c3hj8A_
11.8
17
PDB header: oxidoreductaseChain: A: PDB Molecule: catechol 1,2-dioxygenase;PDBTitle: crystal structure determination of catechol 1,2-dioxygenase from2 rhodococcus opacus 1cp in complex with 4-chlorocatechol
17 d2j0wa1
10.8
47
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like
18 d1zl0a2
9.1
20
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: LD-carboxypeptidase A N-terminal domain-like
19 d1pj3a2
9.1
22
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Malic enzyme N-domain
20 d2cdqa1
9.0
47
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like
21 c4o0lA_
not modelled
8.7
16
PDB header: oxidoreductaseChain: A: PDB Molecule: nadph-dependent 3-quinuclidinone reductase;PDBTitle: crystal structure of nadph-dependent 3-quinuclidinone reductase from2 rhodotorula rubra
22 c3lyhB_
not modelled
8.7
20
PDB header: lyaseChain: B: PDB Molecule: cobalamin (vitamin b12) biosynthesis cbix protein;PDBTitle: crystal structure of putative cobalamin (vitamin b12) biosynthesis2 cbix protein (yp_958415.1) from marinobacter aquaeolei vt8 at 1.60 a3 resolution
23 c3up8B_
not modelled
8.1
19
PDB header: oxidoreductaseChain: B: PDB Molecule: putative 2,5-diketo-d-gluconic acid reductase b;PDBTitle: crystal structure of a putative 2,5-diketo-d-gluconic acid reductase b
24 d1xq1a_
not modelled
8.1
25
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases
25 d1c4qa_
not modelled
8.1
48
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits
26 d1jbja2
not modelled
7.9
40
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains
27 d2bura1
not modelled
7.8
25
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase
28 d1o0sa2
not modelled
7.7
23
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Malic enzyme N-domain
29 d2hmfa1
not modelled
7.2
37
Fold: Carbamate kinase-likeSuperfamily: Carbamate kinase-likeFamily: PyrH-like
30 c5xftA_
not modelled
7.1
22
PDB header: oxidoreductaseChain: A: PDB Molecule: dehydroascorbate reductase;PDBTitle: crystal structure of chlamydomonas reinhardtii dehydroascorbate2 reductase
31 c6hauA_
not modelled
6.4
15
PDB header: rna binding proteinChain: A: PDB Molecule: mrna export factor icp27 homolog;PDBTitle: kshv pan rna mta-response element fragment complexed with the globular2 domain of herpesvirus saimiri orf57
32 c3c1nA_
not modelled
6.3
37
PDB header: transferaseChain: A: PDB Molecule: probable aspartokinase;PDBTitle: crystal structure of allosteric inhibition threonine-sensitive2 aspartokinase from methanococcus jannaschii with l-threonine
33 c2azqA_
not modelled
6.3
26
PDB header: oxidoreductaseChain: A: PDB Molecule: catechol 1,2-dioxygenase;PDBTitle: crystal structure of catechol 1,2-dioxygenase from pseudomonas arvilla2 c-1
34 c2hsiB_
not modelled
6.3
48
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative peptidase m23;PDBTitle: crystal structure of putative peptidase m23 from2 pseudomonas aeruginosa, new york structural genomics3 consortium
35 c1vw46_
not modelled
6.2
14
PDB header: ribosomeChain: 6: PDB Molecule: 54s ribosomal protein l17, mitochondrial;PDBTitle: structure of the yeast mitochondrial large ribosomal subunit
36 d1g0wa1
not modelled
6.2
28
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain
37 c5ou5C_
not modelled
6.2
18
PDB header: photosynthesisChain: C: PDB Molecule: malic enzyme;PDBTitle: crystal structure of maize chloroplastic photosynthetic nadp(+)-2 dependent malic enzyme
38 c2j0wA_
not modelled
5.9
47
PDB header: transferaseChain: A: PDB Molecule: lysine-sensitive aspartokinase 3;PDBTitle: crystal structure of e. coli aspartokinase iii in complex2 with aspartate and adp (r-state)
39 d1zq1c1
not modelled
5.9
35
Fold: GatB/YqeY motifSuperfamily: GatB/YqeY motifFamily: GatB/GatE C-terminal domain-like
40 c5zb3B_
not modelled
5.9
15
PDB header: transcriptionChain: B: PDB Molecule: orf57;PDBTitle: dimeric crystal structure of orf57 from kshv
41 c4z1mJ_
not modelled
5.8
86
PDB header: hydrolaseChain: J: PDB Molecule: atpase inhibitor, mitochondrial;PDBTitle: bovine f1-atpase inhibited by three copies of the inhibitor protein2 if1 crystallised in the presence of thiophosphate.
42 d1s68a_
not modelled
5.6
14
Fold: ATP-graspSuperfamily: DNA ligase/mRNA capping enzyme, catalytic domainFamily: RNA ligase
43 d2jb0b1
not modelled
5.6
36
Fold: His-Me finger endonucleasesSuperfamily: His-Me finger endonucleasesFamily: HNH-motif
44 c2aw5A_
not modelled
5.5
21
PDB header: oxidoreductaseChain: A: PDB Molecule: nadp-dependent malic enzyme;PDBTitle: crystal structure of a human malic enzyme
45 c5vg2B_
not modelled
5.4
17
PDB header: oxidoreductaseChain: B: PDB Molecule: intradiol ring-cleavage dioxygenase;PDBTitle: intradiol ring-cleavage dioxygenase from tetranychus urticae
46 c4uhpA_
not modelled
5.4
24
PDB header: hydrolaseChain: A: PDB Molecule: large component of pyocin ap41;PDBTitle: crystal structure of the pyocin ap41 dnase-immunity complex
47 c3pl0B_
not modelled
5.4
26
PDB header: biosynthetic proteinChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a bsma homolog (mpe_a2762) from methylobium2 petroleophilum pm1 at 1.91 a resolution
48 c3ty8A_
not modelled
5.3
22
PDB header: transferaseChain: A: PDB Molecule: polynucleotide 2',3'-cyclic phosphate phosphodiesterase /PDBTitle: crystal structure of c. thermocellum pnkp ligase domain apo form
49 c4tt3J_
not modelled
5.2
86
PDB header: hydrolaseChain: J: PDB Molecule: atpase inhibitor, mitochondrial;PDBTitle: the pathway of binding of the intrinsically disordered mitochondrial2 inhibitor protein to f1-atpase
50 c1xofA_
not modelled
5.1
100
PDB header: de novo proteinChain: A: PDB Molecule: bbahett1;PDBTitle: heterooligomeric beta beta alpha miniprotein
51 c2jesG_
not modelled
5.1
29
PDB header: viral proteinChain: G: PDB Molecule: portal protein;PDBTitle: portal protein (gp6) from bacteriophage spp1
52 c1fwxB_
not modelled
5.1
21
PDB header: oxidoreductaseChain: B: PDB Molecule: nitrous oxide reductase;PDBTitle: crystal structure of nitrous oxide reductase from p. denitrificans
53 c2boyC_
not modelled
5.1
19
PDB header: oxidoreductaseChain: C: PDB Molecule: 3-chlorocatechol 1,2-dioxygenase;PDBTitle: crystal structure of 3-chlorocatechol 1,2-dioxygenase from rhodococcus2 opacus 1cp