1 c3isrB_
100.0
23
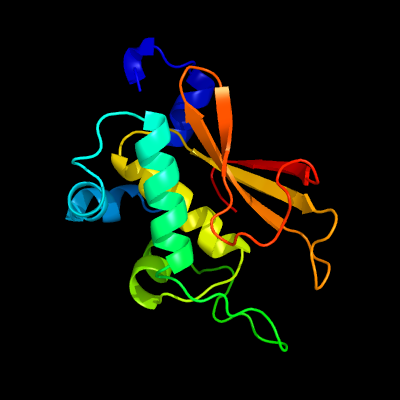
PDB header: hydrolaseChain: B: PDB Molecule: transglutaminase-like enzymes, putative cysteine protease;PDBTitle: the crystal structure of a putative cysteine protease from cytophaga2 hutchinsonii to 1.9a
2 c6g49A_
100.0
18
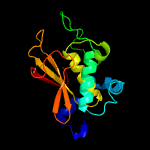
PDB header: transferaseChain: A: PDB Molecule: protein-glutamine gamma-glutamyltransferase;PDBTitle: crystal structure of the periplasmic domain of tgpa from pseudomonas2 aeruginosa
3 c4xz7A_
99.9
21
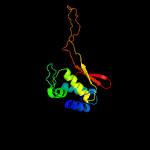
PDB header: transferaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a tgase
4 c3kd4A_
99.7
13
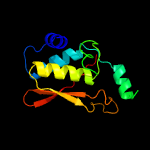
PDB header: hydrolaseChain: A: PDB Molecule: putative protease;PDBTitle: crystal structure of a putative protease (bdi_1141) from2 parabacteroides distasonis atcc 8503 at 2.00 a resolution
5 d2q3za4
99.7
21
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core
6 d1g0da4
99.6
22
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core
7 d2f4ma1
99.6
30
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core
8 d1vjja4
99.6
23
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core
9 d1ex0a4
99.5
27
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core
10 d1x3za1
99.2
20
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core
11 c1kv3F_
99.2
21
PDB header: transferaseChain: F: PDB Molecule: protein-glutamine gamma-glutamyltransferase;PDBTitle: human tissue transglutaminase in gdp bound form
12 c1g0dA_
99.1
22
PDB header: transferaseChain: A: PDB Molecule: protein-glutamine gamma-glutamyltransferase;PDBTitle: crystal structure of red sea bream transglutaminase
13 c1f13A_
99.1
28
PDB header: coagulation factorChain: A: PDB Molecule: cellular coagulation factor xiii zymogen;PDBTitle: recombinant human cellular coagulation factor xiii
14 c1l9mB_
99.1
25
PDB header: transferaseChain: B: PDB Molecule: protein-glutamine glutamyltransferase e3;PDBTitle: three-dimensional structure of the human transglutaminase 32 enzyme: binding of calcium ions change structure for3 activation
15 c4u65F_
99.0
16
PDB header: transferase/hydrolaseChain: F: PDB Molecule: putative cystine protease;PDBTitle: structure of the periplasmic output domain of the legionella2 pneumophila lapd ortholog cdgs9 in complex with pseudomonas3 fluorescens lapg
16 c2qshA_
98.9
12
PDB header: dna binding protein/dnaChain: A: PDB Molecule: dna repair protein rad4;PDBTitle: crystal structure of rad4-rad23 bound to a mismatch dna
17 c3eswA_
98.9
19
PDB header: hydrolaseChain: A: PDB Molecule: peptide-n(4)-(n-acetyl-beta-glucosaminyl)asparaginePDBTitle: complex of yeast pngase with glcnac2-iac.
18 c2pfrB_
98.0
11
PDB header: transferaseChain: B: PDB Molecule: arylamine n-acetyltransferase 2;PDBTitle: human n-acetyltransferase 2
19 c4fgpB_
98.0
13
PDB header: hydrolaseChain: B: PDB Molecule: periplasmic protein;PDBTitle: legionella pneumophila lapg (egta-treated)
20 d1e2ta_
97.9
12
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Arylamine N-acetyltransferase
21 d1w4ta1
not modelled
97.8
12
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Arylamine N-acetyltransferase
22 c3lnbA_
not modelled
97.8
11
PDB header: transferaseChain: A: PDB Molecule: n-acetyltransferase family protein;PDBTitle: crystal structure analysis of arylamine n-acetyltransferase c from2 bacillus anthracis
23 c4guzA_
not modelled
97.7
12
PDB header: transferaseChain: A: PDB Molecule: probable arylamine n-acetyl transferase;PDBTitle: structure of the arylamine n-acetyltransferase from mycobacterium2 abscessus
24 d2bsza1
not modelled
97.6
10
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Arylamine N-acetyltransferase
25 c4dmoB_
not modelled
97.6
9
PDB header: transferaseChain: B: PDB Molecule: n-hydroxyarylamine o-acetyltransferase;PDBTitle: crystal structure of the (baccr)nat3 arylamine n-acetyltransferase2 from bacillus cereus reveals a unique cys-his-glu catalytic triad
26 c2vfbA_
not modelled
97.3
9
PDB header: transferaseChain: A: PDB Molecule: arylamine n-acetyltransferase;PDBTitle: the structure of mycobacterium marinum arylamine n-2 acetyltransferase
27 d1w5ra1
not modelled
97.3
9
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Arylamine N-acetyltransferase
28 c3d9wA_
not modelled
97.2
13
PDB header: transferaseChain: A: PDB Molecule: putative acetyltransferase;PDBTitle: crystal structure analysis of nocardia farcinica arylamine n-2 acetyltransferase
29 d1yuaa2
not modelled
57.1
18
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Prokaryotic DNA topoisomerase I, a C-terminal fragment
30 c1ezaA_
not modelled
48.5
17
PDB header: phosphotransferaseChain: A: PDB Molecule: enzyme i;PDBTitle: amino terminal domain of enzyme i from escherichia coli nmr,2 restrained regularized mean structure
31 c5woyA_
not modelled
47.3
14
PDB header: transferaseChain: A: PDB Molecule: phosphoenolpyruvate-protein phosphotransferase;PDBTitle: nmr solution structure of enzyme i (neit) protein using two 4d-spectra
32 c1yuaA_
not modelled
44.6
17
PDB header: dna binding proteinChain: A: PDB Molecule: topoisomerase i;PDBTitle: c-terminal domain of escherichia coli topoisomerase i
33 c6h8mB_
not modelled
41.7
21
PDB header: hydrolaseChain: B: PDB Molecule: neurotrypsin;PDBTitle: crystal structure of the third srcr domain of murine neurotrypsin.
34 c2jopA_
not modelled
40.0
21
PDB header: immune systemChain: A: PDB Molecule: t-cell surface glycoprotein cd5;PDBTitle: solution structure of the n-terminal extracellular domain2 of the lymphocyte receptor cd5 (cd5 domain 1)
35 d1kbla2
not modelled
36.6
18
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain
36 c4rulA_
not modelled
33.9
25
PDB header: isomerase/dnaChain: A: PDB Molecule: dna topoisomerase 1;PDBTitle: crystal structure of full-length e.coli topoisomerase i in complex2 with ssdna
37 d1h6za2
not modelled
33.6
14
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain
38 c3nrlB_
not modelled
33.1
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein rumgna_01417;PDBTitle: crystal structure of protein rumgna_01417 from ruminococcus gnavus,2 northeast structural genomics consortium target ugr76
39 c2ottY_
not modelled
30.8
15
PDB header: immune systemChain: Y: PDB Molecule: t-cell surface glycoprotein cd5;PDBTitle: crystal structure of cd5_diii
40 c3chgB_
not modelled
29.8
8
PDB header: ligand binding proteinChain: B: PDB Molecule: glycine betaine-binding protein;PDBTitle: the compatible solute-binding protein opuac from bacillus2 subtilis in complex with dmsa
41 d1ou8a_
not modelled
26.5
14
Fold: SspB-likeSuperfamily: SspB-likeFamily: Stringent starvation protein B, SspB
42 c1vbhA_
not modelled
25.6
13
PDB header: transferaseChain: A: PDB Molecule: pyruvate,orthophosphate dikinase;PDBTitle: pyruvate phosphate dikinase with bound mg-pep from maize
43 c3jsyA_
not modelled
25.2
32
PDB header: ribosomal proteinChain: A: PDB Molecule: acidic ribosomal protein p0 homolog;PDBTitle: n-terminal fragment of ribosomal protein l10 from methanococcus2 jannaschii
44 d1zszc1
not modelled
24.4
14
Fold: SspB-likeSuperfamily: SspB-likeFamily: Stringent starvation protein B, SspB
45 d1ou9a_
not modelled
23.4
14
Fold: SspB-likeSuperfamily: SspB-likeFamily: Stringent starvation protein B, SspB
46 c6cgqA_
not modelled
22.3
13
PDB header: lyaseChain: A: PDB Molecule: threonine synthase;PDBTitle: threonine synthase from bacillus subtilis atcc 6633 with plp and plp-2 ala
47 c2a5wC_
not modelled
22.1
19
PDB header: oxidoreductaseChain: C: PDB Molecule: sulfite reductase, desulfoviridin-type subunit gammaPDBTitle: crystal structure of the oxidized gamma-subunit of the dissimilatory2 sulfite reductase (dsrc) from archaeoglobus fulgidus
48 c3j21k_
not modelled
22.1
18
PDB header: ribosomeChain: K: PDB Molecule: 50s ribosomal protein l14e;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
49 d1j0aa_
not modelled
22.0
17
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
50 c3a56B_
not modelled
20.9
12
PDB header: hydrolaseChain: B: PDB Molecule: protein-glutaminase;PDBTitle: crystal structure of pro- protein-glutaminase
51 c4nwbA_
not modelled
20.9
17
PDB header: unknown functionChain: A: PDB Molecule: mrna turnover protein 4;PDBTitle: crystal structure of mrt4
52 c5t1oB_
not modelled
20.5
27
PDB header: transferaseChain: B: PDB Molecule: phosphoenolpyruvate-protein phosphotransferase ptsp;PDBTitle: solution-state nmr and saxs structural ensemble of npr (1-85) in2 complex with ein-ntr (170-424)
53 d1zyma2
not modelled
20.3
25
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: N-terminal domain of enzyme I of the PEP:sugar phosphotransferase system
54 d1yfna1
not modelled
19.6
14
Fold: SspB-likeSuperfamily: SspB-likeFamily: Stringent starvation protein B, SspB
55 d1p5ja_
not modelled
19.6
25
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
56 c1p5jA_
not modelled
19.6
25
PDB header: lyaseChain: A: PDB Molecule: l-serine dehydratase;PDBTitle: crystal structure analysis of human serine dehydratase
57 c2hwgA_
not modelled
19.5
20
PDB header: transferaseChain: A: PDB Molecule: phosphoenolpyruvate-protein phosphotransferase;PDBTitle: structure of phosphorylated enzyme i of the phosphoenolpyruvate:sugar2 phosphotransferase system
58 c1yx3A_
not modelled
19.3
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein dsrc;PDBTitle: nmr structure of allochromatium vinosum dsrc: northeast2 structural genomics consortium target op4
59 c1kblA_
not modelled
18.5
17
PDB header: transferaseChain: A: PDB Molecule: pyruvate phosphate dikinase;PDBTitle: pyruvate phosphate dikinase
60 c3izcs_
not modelled
17.9
53
PDB header: ribosomeChain: S: PDB Molecule: 60s ribosomal protein rpl20 (l18ae);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
61 c2rkbE_
not modelled
17.4
25
PDB header: lyaseChain: E: PDB Molecule: serine dehydratase-like;PDBTitle: serine dehydratase like-1 from human cancer cells
62 d3be7a1
not modelled
15.9
11
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: Zn-dependent arginine carboxypeptidase-like
63 c3iz5s_
not modelled
15.9
18
PDB header: ribosomeChain: S: PDB Molecule: 60s ribosomal protein l18a (l18ae);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
64 c6ei1A_
not modelled
15.6
21
PDB header: hydrolaseChain: A: PDB Molecule: zinc finger with ufm1-specific peptidase domain protein;PDBTitle: crystal structure of the covalent complex between deubiquitinase zufsp2 (zup1) and ubiquitin-pa
65 c2hroA_
not modelled
15.4
23
PDB header: transferaseChain: A: PDB Molecule: phosphoenolpyruvate-protein phosphotransferase;PDBTitle: structure of the full-lenght enzyme i of the pts system from2 staphylococcus carnosus
66 d1vbga2
not modelled
15.3
17
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain
67 c5xa2B_
not modelled
15.2
13
PDB header: transferaseChain: B: PDB Molecule: cysteine synthase;PDBTitle: crystal structure of o-acetylserine sulfhydrylase from planctomyces2 limnophila
68 d1wkva1
not modelled
14.9
25
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
69 c3qp1A_
not modelled
14.6
13
PDB header: transcriptionChain: A: PDB Molecule: cvir transcriptional regulator;PDBTitle: crystal structure of cvir ligand-binding domain bound to the native2 ligand c6-hsl
70 c5xenB_
not modelled
14.3
12
PDB header: transferaseChain: B: PDB Molecule: cysteine synthase;PDBTitle: crystal structure of a hydrogen sulfide-producing enzyme (fn1220) from2 fusobacterium nucleatum in complex with l-serine-plp schiff base
71 c3j3bq_
not modelled
13.8
26
PDB header: ribosomeChain: Q: PDB Molecule: 60s ribosomal protein l18;PDBTitle: structure of the human 60s ribosomal proteins
72 c5b3kA_
not modelled
13.8
24
PDB header: electron transportChain: A: PDB Molecule: uncharacterized protein pa3435;PDBTitle: c101a mutant of flavodoxin from pseudomonas aeruginosa
73 c5ybwA_
not modelled
13.7
12
PDB header: isomeraseChain: A: PDB Molecule: aspartate racemase;PDBTitle: crystal structure of pyridoxal 5'-phosphate-dependent aspartate2 racemase
74 c4d8tC_
not modelled
13.7
12
PDB header: lyaseChain: C: PDB Molecule: d-cysteine desulfhydrase;PDBTitle: crystal structure of d-cysteine desulfhydrase from salmonella2 typhimurium at 2.2 a resolution
75 d1jvaa3
not modelled
13.6
19
Fold: Homing endonuclease-likeSuperfamily: Homing endonucleasesFamily: Intein endonuclease
76 c4hubG_
not modelled
13.4
37
PDB header: ribosomeChain: G: PDB Molecule: 50s ribosomal protein l10e;PDBTitle: the re-refined crystal structure of the haloarcula marismortui large2 ribosomal subunit at 2.4 angstrom resolution: more complete structure3 of the l7/l12 and l1 stalk, l5 and lx proteins
77 c2gn0A_
not modelled
13.1
9
PDB header: lyaseChain: A: PDB Molecule: threonine dehydratase catabolic;PDBTitle: crystal structure of dimeric biodegradative threonine deaminase (tdcb)2 from salmonella typhimurium at 1.7 a resolution (triclinic form with3 one complete subunit built in alternate conformation)
78 c2j0wA_
not modelled
12.6
28
PDB header: transferaseChain: A: PDB Molecule: lysine-sensitive aspartokinase 3;PDBTitle: crystal structure of e. coli aspartokinase iii in complex2 with aspartate and adp (r-state)
79 c2f4qA_
not modelled
12.5
32
PDB header: isomeraseChain: A: PDB Molecule: type i topoisomerase, putative;PDBTitle: crystal structure of deinococcus radiodurans topoisomerase ib
80 c2d1fA_
not modelled
11.9
20
PDB header: lyaseChain: A: PDB Molecule: threonine synthase;PDBTitle: structure of mycobacterium tuberculosis threonine synthase
81 d1fcja_
not modelled
11.9
8
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
82 c2m3dA_
not modelled
11.7
21
PDB header: hydrolaseChain: A: PDB Molecule: nucleolar rna helicase 2;PDBTitle: nmr structure of the guct domain from human dead box polypeptide 21
83 c4airB_
not modelled
11.7
17
PDB header: transferaseChain: B: PDB Molecule: cysteine synthase;PDBTitle: leishmania major cysteine synthase
84 d1tyza_
not modelled
11.6
8
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
85 c2ct6A_
not modelled
11.5
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sh3 domain-binding glutamic acid-rich-likePDBTitle: solution structure of the sh3 domain-binding glutamic acid-2 rich-like protein 2
86 c2zsjB_
not modelled
11.3
24
PDB header: lyaseChain: B: PDB Molecule: threonine synthase;PDBTitle: crystal structure of threonine synthase from aquifex aeolicus vf5
87 d2e29a1
not modelled
11.3
15
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: GUCT domain
88 d1f2da_
not modelled
11.1
13
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
89 c5cvcB_
not modelled
11.1
21
PDB header: isomeraseChain: B: PDB Molecule: serine racemase;PDBTitle: structure of maize serine racemase
90 c4aecB_
not modelled
11.0
4
PDB header: lyaseChain: B: PDB Molecule: cysteine synthase, mitochondrial;PDBTitle: crystal structure of the arabidopsis thaliana o-acetyl-serine-(thiol)-2 lyase c
91 c5c3uA_
not modelled
9.8
20
PDB header: lyaseChain: A: PDB Molecule: l-serine ammonia-lyase;PDBTitle: crystal structure of a fungal l-serine ammonia-lyase from rhizomucor2 miehei
92 d1tdja1
not modelled
9.8
25
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
93 c4ql4A_
not modelled
9.7
0
PDB header: lyaseChain: A: PDB Molecule: o-acetylserine lyase;PDBTitle: crystal structure of o-acetylserine sulfhydrylase from bacillus2 anthracis
94 d1by2a_
not modelled
9.6
14
Fold: SRCR-likeSuperfamily: SRCR-likeFamily: Scavenger receptor cysteine-rich (SRCR) domain
95 c4xvzB_
not modelled
9.5
22
PDB header: transferaseChain: B: PDB Molecule: mycinamicin iii 3''-o-methyltransferase;PDBTitle: mycf mycinamicin iii 3'-o-methyltransferase in complex with mg
96 c5fbtA_
not modelled
9.5
18
PDB header: transferase/antibioticChain: A: PDB Molecule: phosphoenolpyruvate synthase;PDBTitle: crystal structure of rifampin phosphotransferase rph-lm from listeria2 monocytogenes in complex with rifampin
97 c1x1qA_
not modelled
9.5
8
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase beta chain;PDBTitle: crystal structure of tryptophan synthase beta chain from thermus2 thermophilus hb8
98 d1h59b_
not modelled
9.3
19
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Growth factor receptor domainFamily: Growth factor receptor domain
99 c5ancC_
not modelled
9.3
12
PDB header: translationChain: C: PDB Molecule: 60s acidic ribosomal protein p0;PDBTitle: mechanism of eif6 release from the nascent 60s ribosomal subunit