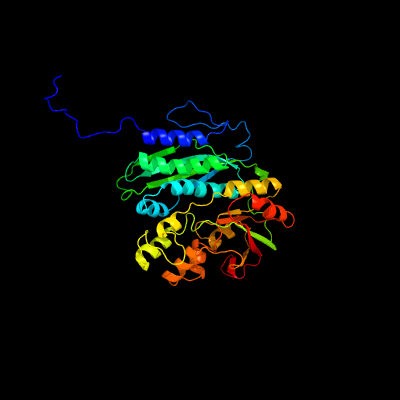
| 1 | c3m84A_
|
|
 |
100.0 |
32 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structure of phosphoribosylaminoimidazole synthetase from2 francisella tularensis
|
|
|
|
| 2 | c3mdoB_
|
|
 |
100.0 |
24 |
PDB header:ligase
Chain: B: PDB Molecule:putative phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structure of a putative phosphoribosylformylglycinamidine2 cyclo-ligase (bdi_2101) from parabacteroides distasonis atcc 8503 at3 1.91 a resolution
|
|
|
|
| 3 | c1cliD_
|
|
 |
100.0 |
45 |
PDB header:ligase
Chain: D: PDB Molecule:protein (phosphoribosyl-aminoimidazole synthetase);
PDBTitle: x-ray crystal structure of aminoimidazole ribonucleotide synthetase2 (purm), from the e. coli purine biosynthetic pathway, at 2.5 a3 resolution
|
|
|
|
| 4 | c3kizA_
|
|
 |
100.0 |
24 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structure of putative phosphoribosylformylglycinamidine cyclo-2 ligase (yp_676759.1) from cytophaga hutchinsonii atcc 33406 at 1.50 a3 resolution
|
|
|
|
| 5 | c2z01A_
|
|
 |
100.0 |
45 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structure of phosphoribosylaminoimidazole synthetase from2 geobacillus kaustophilus
|
|
|
|
| 6 | c5vk4B_
|
|
 |
100.0 |
41 |
PDB header:ligase
Chain: B: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structure of a phosphoribosylformylglycinamidine cyclo-ligase2 from neisseria gonorrhoeae bound to amppnp and magnesium
|
|
|
|
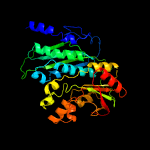
| 7 | c2btuB_
|
|
 |
100.0 |
46 |
PDB header:synthase
Chain: B: PDB Molecule:phosphoribosyl-aminoimidazole synthetase;
PDBTitle: crystal structure of phosphoribosylformylglycinamidine cyclo-ligase2 from bacillus anthracis at 2.3a resolution.
|
|
|
|
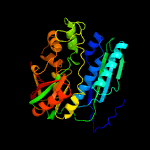
| 8 | c2v9yA_
|
|
 |
100.0 |
45 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: human aminoimidazole ribonucleotide synthetase
|
|
|
|
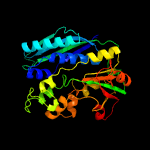
| 9 | c5avmE_
|
|
 |
100.0 |
45 |
PDB header:ligase
Chain: E: PDB Molecule:phosphoribosylformylglycinamidine cyclo-ligase;
PDBTitle: crystal structures of 5-aminoimidazole ribonucleotide (air)2 synthetase, purm, from thermus thermophilus
|
|
|
|
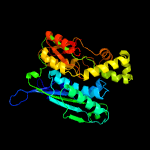
| 10 | c3fd5B_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:selenide, water dikinase 1;
PDBTitle: crystal structure of human selenophosphate synthetase 12 complex with ampcp
|
|
|
|
| 11 | c3mcqA_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:thiamine-monophosphate kinase;
PDBTitle: crystal structure of thiamine-monophosphate kinase (mfla_0573) from2 methylobacillus flagellatus kt at 1.91 a resolution
|
|
|
|
| 12 | c3u0oA_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:selenide, water dikinase;
PDBTitle: the crystal structure of selenophosphate synthetase from e. coli
|
|
|
|
| 13 | c3c9uB_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:thiamine monophosphate kinase;
PDBTitle: aathil complexed with adp and tpp
|
|
|
|
| 14 | c2zauB_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:selenide, water dikinase;
PDBTitle: crystal structure of an n-terminally truncated2 selenophosphate synthetase from aquifex aeolicus
|
|
|
|
| 15 | c3vysC_
|
|
 |
100.0 |
17 |
PDB header:metal binding protein/transferase
Chain: C: PDB Molecule:hydrogenase expression/formation protein hype;
PDBTitle: crystal structure of the hypc-hypd-hype complex (form i)
|
|
|
|
| 16 | c2rb9D_
|
|
 |
100.0 |
21 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:hype protein;
PDBTitle: crystal structure of e.coli hype
|
|
|
|
| 17 | c2zodB_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:selenide, water dikinase;
PDBTitle: crystal structure of selenophosphate synthetase from aquifex aeolicus
|
|
|
|
| 18 | c5cm7A_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:thiamine-monophosphate kinase;
PDBTitle: structure of thiamine-monophosphate kinase from acinetobacter2 baumannii in complex with adenosine diphosphate (adp) and thiamine3 diphosphate (tpp)
|
|
|
|
| 19 | c2z1tA_
|
|
 |
100.0 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:hydrogenase expression/formation protein hype;
PDBTitle: crystal structure of hydrogenase maturation protein hype
|
|
|
|
| 20 | c2yxzA_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:thiamin-monophosphate kinase;
PDBTitle: crystal structure of tt0281 from thermus thermophilus hb8
|
|
|
|
| 21 | c3vtiD_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: D: PDB Molecule:hydrogenase maturation factor;
PDBTitle: crystal structure of hype-hypf complex
|
|
|
| 22 | c1vqvB_ |
|
not modelled |
100.0 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:thiamine monophosphate kinase;
PDBTitle: crystal structure of thiamine monophosphate kinase (thil)2 from aquifex aeolicus
|
|
|
| 23 | c2z1eA_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:hydrogenase expression/formation protein hype;
PDBTitle: crystal structure of hype from thermococcus kodakaraensis (outward2 form)
|
|
|
| 24 | c3ac6A_ |
|
not modelled |
100.0 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine synthase 2;
PDBTitle: crystal structure of purl from thermus thermophilus
|
|
|
| 25 | c2hs0A_ |
|
not modelled |
100.0 |
23 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine synthase ii;
PDBTitle: t. maritima purl complexed with atp
|
|
|
| 26 | d1clia1 |
|
not modelled |
100.0 |
52 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 27 | c5l16A_ |
|
not modelled |
100.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:putative selenophosphate synthetase;
PDBTitle: crystal structure of n-terminus truncated selenophosphate synthetase2 from leishmania major
|
|
|
| 28 | c3d54I_ |
|
not modelled |
100.0 |
22 |
PDB header:ligase
Chain: I: PDB Molecule:phosphoribosylformylglycinamidine synthase ii;
PDBTitle: structure of purlqs from thermotoga maritima
|
|
|
| 29 | d1clib1 |
|
not modelled |
100.0 |
52 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 30 | d1clia2 |
|
not modelled |
100.0 |
39 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 31 | c1t3tA_ |
|
not modelled |
100.0 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylformylglycinamidine synthase;
PDBTitle: structure of formylglycinamide synthetase
|
|
|
| 32 | d3c9ua1 |
|
not modelled |
99.9 |
19 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 33 | d2zoda2 |
|
not modelled |
99.9 |
15 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 34 | d2zaua1 |
|
not modelled |
99.9 |
19 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 35 | d2z1ea2 |
|
not modelled |
99.9 |
16 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 36 | d3c9ua2 |
|
not modelled |
99.9 |
18 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 37 | d2z1ea1 |
|
not modelled |
99.9 |
20 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 38 | d2zoda1 |
|
not modelled |
99.9 |
19 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 39 | d1vk3a2 |
|
not modelled |
99.9 |
17 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 40 | d1vk3a1 |
|
not modelled |
99.8 |
26 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 41 | d1vk3a3 |
|
not modelled |
99.8 |
22 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 42 | d1t3ta6 |
|
not modelled |
99.7 |
13 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 43 | d1t3ta7 |
|
not modelled |
99.6 |
18 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
|
|
| 44 | d1t3ta4 |
|
not modelled |
99.2 |
19 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 45 | d1t3ta5 |
|
not modelled |
98.2 |
12 |
Fold:Bacillus chorismate mutase-like
Superfamily:PurM N-terminal domain-like
Family:PurM N-terminal domain-like |
|
|
| 46 | d1to3a_ |
|
not modelled |
81.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 47 | d3dhxa1 |
|
not modelled |
76.6 |
32 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:NIL domain-like |
|
|
| 48 | c5kdmD_ |
|
not modelled |
74.2 |
13 |
PDB header:chaperone / dna binding protein
Chain: D: PDB Molecule:major tegument protein;
PDBTitle: crystal structure of ebv tegument protein bnrf1 in complex with2 histone chaperone daxx and histones h3.3-h4
|
|
|
| 49 | c3dzvB_ |
|
not modelled |
66.7 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:4-methyl-5-(beta-hydroxyethyl)thiazole kinase;
PDBTitle: crystal structure of 4-methyl-5-(beta-hydroxyethyl)thiazole kinase2 (np_816404.1) from enterococcus faecalis v583 at 2.57 a resolution
|
|
|
| 50 | c5cgaC_ |
|
not modelled |
58.5 |
27 |
PDB header:transferase
Chain: C: PDB Molecule:hydroxyethylthiazole kinase;
PDBTitle: structure of hydroxyethylthiazole kinase thim from staphylococcus2 aureus in complex with substrate analog 2-(1,3,5-trimethyl-1h-3 pyrazole-4-yl)ethanol
|
|
|
| 51 | d1q3oa_ |
|
not modelled |
54.6 |
15 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
|
|
| 52 | d2qrra1 |
|
not modelled |
50.3 |
23 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:NIL domain-like |
|
|
| 53 | c3l4fD_ |
|
not modelled |
48.5 |
15 |
PDB header:signaling protein/protein binding
Chain: D: PDB Molecule:sh3 and multiple ankyrin repeat domains protein 1;
PDBTitle: crystal structure of betapix coiled-coil domain and shank pdz complex
|
|
|
| 54 | c2q6tB_ |
|
not modelled |
47.2 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:dnab replication fork helicase;
PDBTitle: crystal structure of the thermus aquaticus dnab monomer
|
|
|
| 55 | c3qglD_ |
|
not modelled |
40.5 |
12 |
PDB header:protein binding
Chain: D: PDB Molecule:sorting nexin-27;
PDBTitle: crystal structure of pdz domain of sorting nexin 27 (snx27) in complex2 with the eseskv peptide corresponding to the c-terminal tail of girk3
|
|
|
| 56 | c3jrkG_ |
|
not modelled |
38.9 |
15 |
PDB header:lyase
Chain: G: PDB Molecule:tagatose 1,6-diphosphate aldolase 2;
PDBTitle: a putative tagatose 1,6-diphosphate aldolase from streptococcus2 pyogenes
|
|
|
| 57 | d1v8aa_ |
|
not modelled |
37.4 |
20 |
Fold:Ribokinase-like
Superfamily:Ribokinase-like
Family:Thiamin biosynthesis kinases |
|
|
| 58 | c5f67A_ |
|
not modelled |
36.2 |
20 |
PDB header:protein binding
Chain: A: PDB Molecule:inactivation-no-after-potential d protein;
PDBTitle: an exquisitely specific pdz/target recognition revealed by the2 structure of inad pdz3 in complex with trp channel tail
|
|
|
| 59 | d1jyea_ |
|
not modelled |
35.2 |
26 |
Fold:Periplasmic binding protein-like I
Superfamily:Periplasmic binding protein-like I
Family:L-arabinose binding protein-like |
|
|
| 60 | c1jyeA_ |
|
not modelled |
35.2 |
26 |
PDB header:transcription
Chain: A: PDB Molecule:lactose operon repressor;
PDBTitle: structure of a dimeric lac repressor with c-terminal deletion and k84l2 substitution
|
|
|
| 61 | c4a1fB_ |
|
not modelled |
34.7 |
8 |
PDB header:hydrolase
Chain: B: PDB Molecule:replicative dna helicase;
PDBTitle: crystal structure of c-terminal domain of helicobacter2 pylori dnab helicase
|
|
|
| 62 | c2xkxB_ |
|
not modelled |
34.2 |
18 |
PDB header:structural protein
Chain: B: PDB Molecule:disks large homolog 4;
PDBTitle: single particle analysis of psd-95 in negative stain
|
|
|
| 63 | c1vjqB_ |
|
not modelled |
33.6 |
22 |
PDB header:structural genomics, de novo protein
Chain: B: PDB Molecule:designed protein;
PDBTitle: designed protein based on backbone conformation of2 procarboxypeptidase-a (1aye) with sidechains chosen for maximal3 predicted stability.
|
|
|
| 64 | d1n55a_ |
|
not modelled |
32.1 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 65 | c3qstA_ |
|
not modelled |
32.0 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase, putative;
PDBTitle: crystal structure of trichomonas vaginalis triosephosphate isomerase2 tvag_096350 gene (val-45 variant)
|
|
|
| 66 | d2csua1 |
|
not modelled |
31.8 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
| 67 | c6hyhA_ |
|
not modelled |
31.5 |
20 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:periplasmic binding protein/laci transcriptional regulator;
PDBTitle: crystal structure of msmeg_1712 from mycobacterium smegmatis in2 complex with beta-d-fucofuranose
|
|
|
| 68 | c3shwA_ |
|
not modelled |
31.4 |
18 |
PDB header:cell adhesion
Chain: A: PDB Molecule:tight junction protein zo-1;
PDBTitle: crystal structure of zo-1 pdz3-sh3-guk supramodule complex with2 connexin-45 peptide
|
|
|
| 69 | c3d8uA_ |
|
not modelled |
30.8 |
12 |
PDB header:transcription regulator
Chain: A: PDB Molecule:purr transcriptional regulator;
PDBTitle: the crystal structure of a purr family transcriptional regulator from2 vibrio parahaemolyticus rimd 2210633
|
|
|
| 70 | d1trea_ |
|
not modelled |
30.2 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 71 | c4ohqB_ |
|
not modelled |
30.2 |
20 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase, chloroplastic;
PDBTitle: crystal structure of chloroplast triose phosphate isomerase from2 arabidopsis thaliana
|
|
|
| 72 | c5uprA_ |
|
not modelled |
29.8 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: x-ray structure of a putative triosephosphate isomerase from2 toxoplasma gondii me49
|
|
|
| 73 | d2btma_ |
|
not modelled |
29.2 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 74 | c4nmnA_ |
|
not modelled |
29.1 |
9 |
PDB header:replication
Chain: A: PDB Molecule:replicative dna helicase;
PDBTitle: aquifex aeolicus replicative helicase (dnab) complexed with adp, at2 3.3 resolution
|
|
|
| 75 | c4y8fA_ |
|
not modelled |
29.0 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from clostridium2 perfringens
|
|
|
| 76 | c3bgwD_ |
|
not modelled |
28.0 |
7 |
PDB header:replication
Chain: D: PDB Molecule:dnab-like replicative helicase;
PDBTitle: the structure of a dnab-like replicative helicase and its interactions2 with primase
|
|
|
| 77 | c2vk2A_ |
|
not modelled |
27.1 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:abc transporter periplasmic-binding protein ytfq;
PDBTitle: crystal structure of a galactofuranose binding protein
|
|
|
| 78 | c3rotA_ |
|
not modelled |
27.0 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:abc sugar transporter, periplasmic sugar binding protein;
PDBTitle: crystal structure of abc sugar transporter (periplasmic sugar binding2 protein) from legionella pneumophila
|
|
|
| 79 | d1kv5a_ |
|
not modelled |
27.0 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 80 | c4y9aB_ |
|
not modelled |
26.1 |
22 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from streptomyces2 coelicolor
|
|
|
| 81 | c6c8rA_ |
|
not modelled |
25.7 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:loganic acid o-methyltransferase;
PDBTitle: loganic acid o-methyltransferase complexed with sah and loganic acid
|
|
|
| 82 | c4zc0A_ |
|
not modelled |
25.4 |
7 |
PDB header:hydrolase
Chain: A: PDB Molecule:replicative dna helicase;
PDBTitle: structure of a dodecameric bacterial helicase
|
|
|
| 83 | d2f0aa1 |
|
not modelled |
25.3 |
16 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
|
|
| 84 | d1m6ja_ |
|
not modelled |
25.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 85 | c5jxfA_ |
|
not modelled |
24.8 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:asp/glu-specific dipeptidyl-peptidase;
PDBTitle: crystal structure of flavobacterium psychrophilum dpp11 in complex2 with dipeptide arg-asp
|
|
|
| 86 | d1suxa_ |
|
not modelled |
24.6 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 87 | c2eg5C_ |
|
not modelled |
24.5 |
32 |
PDB header:transferase
Chain: C: PDB Molecule:xanthosine methyltransferase;
PDBTitle: the structure of xanthosine methyltransferase
|
|
|
| 88 | d1m6ex_ |
|
not modelled |
23.8 |
20 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Salicylic acid carboxyl methyltransferase (SAMT) |
|
|
| 89 | c4rxmA_ |
|
not modelled |
23.7 |
6 |
PDB header:transport protein
Chain: A: PDB Molecule:possible sugar abc superfamily atp binding cassette
PDBTitle: crystal structure of periplasmic abc transporter solute binding2 protein a7jw62 from mannheimia haemolytica phl213, target efi-511105,3 in complex with myo-inositol
|
|
|
| 90 | c4y90B_ |
|
not modelled |
23.5 |
24 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from deinococcus2 radiodurans
|
|
|
| 91 | d1wfga_ |
|
not modelled |
22.9 |
20 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
|
|
| 92 | c5ujwD_ |
|
not modelled |
22.5 |
17 |
PDB header:isomerase
Chain: D: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from francisella2 tularensis subsp. tularensis schu s4
|
|
|
| 93 | c3fokH_ |
|
not modelled |
22.2 |
15 |
PDB header:structural genomics, unknown function
Chain: H: PDB Molecule:uncharacterized protein cgl0159;
PDBTitle: crystal structure of cgl0159 from corynebacterium2 glutamicum (brevibacterium flavum). northeast structural3 genomics target cgr115
|
|
|
| 94 | d1o5xa_ |
|
not modelled |
22.1 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 95 | c3l49D_ |
|
not modelled |
22.0 |
9 |
PDB header:transport protein
Chain: D: PDB Molecule:abc sugar (ribose) transporter, periplasmic substrate-
PDBTitle: crystal structure of abc sugar transporter subunit from rhodobacter2 sphaeroides 2.4.1
|
|
|
| 96 | c4ry8D_ |
|
not modelled |
21.9 |
15 |
PDB header:transport protein
Chain: D: PDB Molecule:periplasmic binding protein;
PDBTitle: crystal structure of 5-methylthioribose transporter solute binding2 protein tlet_1677 from thermotoga lettingae tmo target efi-511109 in3 complex with 5-methylthioribose
|
|
|
| 97 | c2m0tA_ |
|
not modelled |
21.6 |
15 |
PDB header:protein binding
Chain: A: PDB Molecule:na(+)/h(+) exchange regulatory cofactor nhe-rf1;
PDBTitle: structural characterization of the extended pdz1 domain from nherf1
|
|
|
| 98 | c3gndC_ |
|
not modelled |
21.5 |
15 |
PDB header:lyase
Chain: C: PDB Molecule:aldolase lsrf;
PDBTitle: crystal structure of e. coli lsrf in complex with ribulose-5-phosphate
|
|
|
| 99 | c5ocpA_ |
|
not modelled |
21.5 |
6 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:periplasmic binding protein/laci transcriptional regulator;
PDBTitle: the periplasmic binding protein component of the arabinose abc2 transporter from shewanella sp. ana-3 bound to alpha and beta-l-3 arabinofuranose
|
|
|
| 100 | c5braA_ |
|
not modelled |
21.3 |
10 |
PDB header:solute-binding protein
Chain: A: PDB Molecule:putative periplasmic binding protein with substrate ribose;
PDBTitle: crystal structure of a putative periplasmic solute binding protein2 (ipr025997) from ochrobactrum anthropi atcc49188 (oant_2843, target3 efi-511085)
|
|
|
| 101 | c3io5B_ |
|
not modelled |
21.3 |
6 |
PDB header:dna binding protein
Chain: B: PDB Molecule:recombination and repair protein;
PDBTitle: crystal structure of a dimeric form of the uvsx recombinase core2 domain from enterobacteria phage t4
|
|
|
| 102 | c4x22A_ |
|
not modelled |
21.2 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of leptospira interrogans triosephosphate isomerase2 (litim)
|
|
|
| 103 | c4y96B_ |
|
not modelled |
21.1 |
22 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from gemmata2 obscuriglobus
|
|
|
| 104 | c5hsgA_ |
|
not modelled |
21.1 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:putative abc transporter, nucleotide binding/atpase
PDBTitle: crystal structure of an abc transporter solute binding protein from2 klebsiella pneumoniae (kpn_01730, target efi-511059), apo open3 structure
|
|
|
| 105 | c2vyeA_ |
|
not modelled |
21.0 |
9 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:replicative dna helicase;
PDBTitle: crystal structure of the dnac-ssdna complex
|
|
|
| 106 | c1yyaA_ |
|
not modelled |
20.7 |
25 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of tt0473, putative triosephosphate isomerase from2 thermus thermophilus hb8
|
|
|
| 107 | d1pcaa1 |
|
not modelled |
20.7 |
12 |
Fold:Ferredoxin-like
Superfamily:Protease propeptides/inhibitors
Family:Pancreatic carboxypeptidase, activation domain |
|
|
| 108 | c4kzkA_ |
|
not modelled |
20.6 |
6 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:l-arabinose abc transporter, periplasmic l-arabinose-
PDBTitle: the structure of the periplasmic l-arabinose binding protein from2 burkholderia thailandensis
|
|
|
| 109 | d1o6xa_ |
|
not modelled |
20.6 |
8 |
Fold:Ferredoxin-like
Superfamily:Protease propeptides/inhibitors
Family:Pancreatic carboxypeptidase, activation domain |
|
|
| 110 | c4g1kB_ |
|
not modelled |
20.5 |
24 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from burkholderia2 thailandensis
|
|
|
| 111 | c3e61A_ |
|
not modelled |
20.5 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative transcriptional repressor of ribose operon;
PDBTitle: crystal structure of a putative transcriptional repressor of ribose2 operon from staphylococcus saprophyticus subsp. saprophyticus
|
|
|
| 112 | c3jy6B_ |
|
not modelled |
20.4 |
18 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, laci family;
PDBTitle: crystal structure of laci transcriptional regulator from lactobacillus2 brevis
|
|
|
| 113 | c3krsB_ |
|
not modelled |
20.3 |
24 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: structure of triosephosphate isomerase from cryptosporidium parvum at2 1.55a resolution
|
|
|