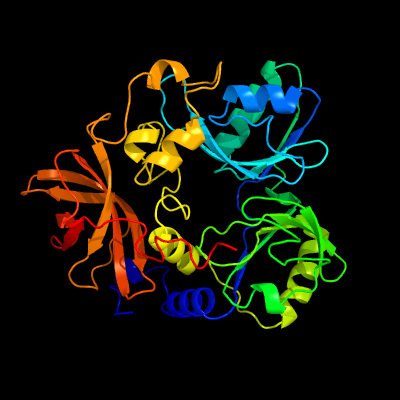
| 1 | c1pj6A_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n,n-dimethylglycine oxidase;
PDBTitle: crystal structure of dimethylglycine oxidase of arthrobacter2 globiformis in complex with folic acid
|
|
|
|
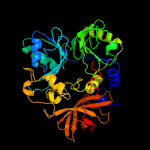
| 2 | c4p9sA_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dimethylglycine dehydrogenase;
PDBTitle: crystal structure of the mature form of rat dmgdh
|
|
|
|
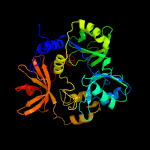
| 3 | c3girA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:aminomethyltransferase;
PDBTitle: crystal structure of glycine cleavage system2 aminomethyltransferase t from bartonella henselae
|
|
|
|
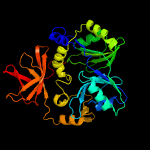
| 4 | c1worA_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:aminomethyltransferase;
PDBTitle: crystal structure of t-protein of the glycine cleavage2 system
|
|
|
|
| 5 | c1x31A_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sarcosine oxidase alpha subunit;
PDBTitle: crystal structure of heterotetrameric sarcosine oxidase from2 corynebacterium sp. u-96
|
|
|
|
| 6 | c1vloA_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:aminomethyltransferase;
PDBTitle: crystal structure of aminomethyltransferase (t protein;2 tetrahydrofolate-dependent) of glycine cleavage system (np417381)3 from escherichia coli k12 at 1.70 a resolution
|
|
|
|
| 7 | c1v5vA_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:aminomethyltransferase;
PDBTitle: crystal structure of a component of glycine cleavage system: t-protein2 from pyrococcus horikoshii ot3 at 1.5 a resolution
|
|
|
|
| 8 | c1wsrA_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:aminomethyltransferase;
PDBTitle: crystal structure of human t-protein of glycine cleavage2 system
|
|
|
|
| 9 | c1yx2B_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:aminomethyltransferase;
PDBTitle: crystal structure of the probable aminomethyltransferase2 from bacillus subtilis
|
|
|
|
| 10 | c3tfhB_
|
|
 |
100.0 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:gcvt-like aminomethyltransferase protein;
PDBTitle: dmsp-dependent demethylase from p. ubique - apo
|
|
|
|
| 11 | c5tl4C_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:vanillate/3-o-methylgallate o-demethylase;
PDBTitle: crystal structure of sphingomonas paucimobilis aryl o-demethylase ligm
|
|
|
|
| 12 | c1vlyA_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:unknown protein from 2d-page;
PDBTitle: crystal structure of a putative aminomethyltransferase (ygfz) from2 escherichia coli at 1.30 a resolution
|
|
|
|
| 13 | c5oliA_
|
|
 |
100.0 |
21 |
PDB header:protein binding
Chain: A: PDB Molecule:putative transferase caf17, mitochondrial;
PDBTitle: crystal structure of human iba57
|
|
|
|
| 14 | d1vloa2
|
|
 |
100.0 |
20 |
Fold:Folate-binding domain
Superfamily:Folate-binding domain
Family:Aminomethyltransferase folate-binding domain |
|
|
|
| 15 | d1wosa2
|
|
 |
100.0 |
19 |
Fold:Folate-binding domain
Superfamily:Folate-binding domain
Family:Aminomethyltransferase folate-binding domain |
|
|
|
| 16 | d1pj5a4
|
|
 |
100.0 |
20 |
Fold:Folate-binding domain
Superfamily:Folate-binding domain
Family:Aminomethyltransferase folate-binding domain |
|
|
|
| 17 | d1v5va2
|
|
 |
100.0 |
18 |
Fold:Folate-binding domain
Superfamily:Folate-binding domain
Family:Aminomethyltransferase folate-binding domain |
|
|
|
| 18 | d1vlya2
|
|
 |
100.0 |
18 |
Fold:Folate-binding domain
Superfamily:Folate-binding domain
Family:Aminomethyltransferase folate-binding domain |
|
|
|
| 19 | c2gagC_
|
|
 |
99.9 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:heterotetrameric sarcosine oxidase gamma-subunit;
PDBTitle: heteroteterameric sarcosine: structure of a diflavin metaloenzyme at2 1.85 a resolution
|
|
|
|
| 20 | d1pj5a1
|
|
 |
99.4 |
17 |
Fold:Elongation factor/aminomethyltransferase common domain
Superfamily:Aminomethyltransferase beta-barrel domain
Family:Aminomethyltransferase beta-barrel domain |
|
|
|
| 21 | d1wosa1 |
|
not modelled |
99.2 |
24 |
Fold:Elongation factor/aminomethyltransferase common domain
Superfamily:Aminomethyltransferase beta-barrel domain
Family:Aminomethyltransferase beta-barrel domain |
|
|
| 22 | c6ah3B_ |
|
not modelled |
99.2 |
18 |
PDB header:hydrolase/rna
Chain: B: PDB Molecule:ribonucleases p/mrp protein subunit pop1;
PDBTitle: cryo-em structure of yeast ribonuclease p with pre-trna substrate
|
|
|
| 23 | d1v5va1 |
|
not modelled |
99.2 |
17 |
Fold:Elongation factor/aminomethyltransferase common domain
Superfamily:Aminomethyltransferase beta-barrel domain
Family:Aminomethyltransferase beta-barrel domain |
|
|
| 24 | c6ahrB_ |
|
not modelled |
99.1 |
23 |
PDB header:hydrolase/rna
Chain: B: PDB Molecule:ribonucleases p/mrp protein subunit pop1;
PDBTitle: cryo-em structure of human ribonuclease p
|
|
|
| 25 | d1vloa1 |
|
not modelled |
99.1 |
20 |
Fold:Elongation factor/aminomethyltransferase common domain
Superfamily:Aminomethyltransferase beta-barrel domain
Family:Aminomethyltransferase beta-barrel domain |
|
|
| 26 | d1vlya1 |
|
not modelled |
97.2 |
20 |
Fold:Elongation factor/aminomethyltransferase common domain
Superfamily:Aminomethyltransferase beta-barrel domain
Family:Aminomethyltransferase beta-barrel domain |
|
|
| 27 | c1xzqA_ |
|
not modelled |
93.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable trna modification gtpase trme;
PDBTitle: structure of the gtp-binding protein trme from thermotoga2 maritima complexed with 5-formyl-thf
|
|
|
| 28 | c3gehA_ |
|
not modelled |
92.8 |
5 |
PDB header:hydrolase
Chain: A: PDB Molecule:trna modification gtpase mnme;
PDBTitle: crystal structure of mnme from nostoc in complex with gdp, folinic2 acid and zn
|
|
|
| 29 | d1xzpa3 |
|
not modelled |
92.6 |
15 |
Fold:Folate-binding domain
Superfamily:Folate-binding domain
Family:TrmE formyl-THF-binding domain |
|
|
| 30 | c1xzqB_ |
|
not modelled |
92.6 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable trna modification gtpase trme;
PDBTitle: structure of the gtp-binding protein trme from thermotoga2 maritima complexed with 5-formyl-thf
|
|
|
| 31 | c3geiB_ |
|
not modelled |
91.9 |
9 |
PDB header:hydrolase
Chain: B: PDB Molecule:trna modification gtpase mnme;
PDBTitle: crystal structure of mnme from chlorobium tepidum in complex2 with gcp
|
|
|
| 32 | c2i5nH_ |
|
not modelled |
35.5 |
18 |
PDB header:photosynthesis
Chain: H: PDB Molecule:reaction center protein h chain;
PDBTitle: 1.96 a x-ray structure of photosynthetic reaction center from2 rhodopseudomonas viridis:crystals grown by microfluidic technique
|
|
|
| 33 | d1eysh1 |
|
not modelled |
32.2 |
13 |
Fold:PRC-barrel domain
Superfamily:PRC-barrel domain
Family:Photosynthetic reaction centre, H-chain, cytoplasmic domain |
|
|
| 34 | c1eysH_ |
|
not modelled |
28.7 |
13 |
PDB header:electron transport
Chain: H: PDB Molecule:photosynthetic reaction center;
PDBTitle: crystal structure of photosynthetic reaction center from a2 thermophilic bacterium, thermochromatium tepidum
|
|
|
| 35 | c5uw8C_ |
|
not modelled |
28.0 |
19 |
PDB header:transport protein
Chain: C: PDB Molecule:probable phospholipid abc transporter-binding protein mlad;
PDBTitle: structure of e. coli mce protein mlad, core mce domain
|
|
|
| 36 | c3l48B_ |
|
not modelled |
26.5 |
24 |
PDB header:transport protein
Chain: B: PDB Molecule:outer membrane usher protein papc;
PDBTitle: crystal structure of the c-terminal domain of the papc usher
|
|
|
| 37 | c5uvnE_ |
|
not modelled |
23.2 |
15 |
PDB header:transport protein
Chain: E: PDB Molecule:paraquat-inducible protein b;
PDBTitle: structure of e. coli mce protein pqib, periplasmic domain
|
|
|
| 38 | c5uvnB_ |
|
not modelled |
23.2 |
15 |
PDB header:transport protein
Chain: B: PDB Molecule:paraquat-inducible protein b;
PDBTitle: structure of e. coli mce protein pqib, periplasmic domain
|
|
|
| 39 | c5uvnA_ |
|
not modelled |
23.2 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:paraquat-inducible protein b;
PDBTitle: structure of e. coli mce protein pqib, periplasmic domain
|
|
|
| 40 | c5uvnC_ |
|
not modelled |
23.2 |
15 |
PDB header:transport protein
Chain: C: PDB Molecule:paraquat-inducible protein b;
PDBTitle: structure of e. coli mce protein pqib, periplasmic domain
|
|
|
| 41 | c5uvnF_ |
|
not modelled |
23.2 |
15 |
PDB header:transport protein
Chain: F: PDB Molecule:paraquat-inducible protein b;
PDBTitle: structure of e. coli mce protein pqib, periplasmic domain
|
|
|
| 42 | c5uvnD_ |
|
not modelled |
23.2 |
15 |
PDB header:transport protein
Chain: D: PDB Molecule:paraquat-inducible protein b;
PDBTitle: structure of e. coli mce protein pqib, periplasmic domain
|
|
|
| 43 | d1w4ma_ |
|
not modelled |
19.9 |
11 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:DEP domain |
|
|
| 44 | d2i5nh1 |
|
not modelled |
19.0 |
18 |
Fold:PRC-barrel domain
Superfamily:PRC-barrel domain
Family:Photosynthetic reaction centre, H-chain, cytoplasmic domain |
|
|
| 45 | d3bzka5 |
|
not modelled |
16.1 |
16 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Tex RuvX-like domain-like |
|
|
| 46 | d1rzhh1 |
|
not modelled |
15.7 |
9 |
Fold:PRC-barrel domain
Superfamily:PRC-barrel domain
Family:Photosynthetic reaction centre, H-chain, cytoplasmic domain |
|
|
| 47 | c2ysrA_ |
|
not modelled |
15.4 |
30 |
PDB header:signaling protein
Chain: A: PDB Molecule:dep domain-containing protein 1;
PDBTitle: solution structure of the dep domain from human dep domain-2 containing protein 1
|
|
|
| 48 | c6ic4C_ |
|
not modelled |
11.7 |
16 |
PDB header:protein transport
Chain: C: PDB Molecule:toluene tolerance efflux transporter (abc superfamily,
PDBTitle: cryo-em structure of the a. baumannii mla complex at 8.7 a resolution
|
|
|
| 49 | d1uhwa_ |
|
not modelled |
10.5 |
9 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:DEP domain |
|
|
| 50 | c5d5pC_ |
|
not modelled |
9.5 |
28 |
PDB header:transferase
Chain: C: PDB Molecule:hcgb;
PDBTitle: hcgb from methanococcus maripaludis
|
|
|
| 51 | d1fsha_ |
|
not modelled |
9.2 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:DEP domain |
|
|
| 52 | d2hjsa1 |
|
not modelled |
7.8 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 53 | c3brcA_ |
|
not modelled |
7.4 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved protein of unknown function;
PDBTitle: crystal structure of a conserved protein of unknown function from2 methanobacterium thermoautotrophicum
|
|
|
| 54 | c3zbqA_ |
|
not modelled |
7.1 |
8 |
PDB header:viral protein
Chain: A: PDB Molecule:phikz039;
PDBTitle: protofilament of tubz from bacteriophage phikz
|
|
|
| 55 | d1rmva_ |
|
not modelled |
6.9 |
14 |
Fold:Four-helical up-and-down bundle
Superfamily:TMV-like viral coat proteins
Family:TMV-like viral coat proteins |
|
|
| 56 | c3kmlB_ |
|
not modelled |
6.7 |
10 |
PDB header:viral protein
Chain: B: PDB Molecule:coat protein;
PDBTitle: circular permutant of the tobacco mosaic virus
|
|
|
| 57 | d2csoa1 |
|
not modelled |
6.6 |
9 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:DEP domain |
|
|
| 58 | c5oqmh_ |
|
not modelled |
6.5 |
28 |
PDB header:transcription
Chain: H: PDB Molecule:dna-directed rna polymerases i, ii, and iii subunit rpabc3;
PDBTitle: structure of yeast transcription pre-initiation complex with tfiih and2 core mediator
|
|
|
| 59 | d1ukfa_ |
|
not modelled |
6.4 |
7 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Avirulence protein Avrpph3 |
|
|
| 60 | c3jc6E_ |
|
not modelled |
6.4 |
11 |
PDB header:replication
Chain: E: PDB Molecule:cell division control protein 45;
PDBTitle: structure of the eukaryotic replicative cmg helicase and pumpjack2 motion
|
|
|
| 61 | d1vqod1 |
|
not modelled |
6.2 |
15 |
Fold:RL5-like
Superfamily:RL5-like
Family:Ribosomal protein L5 |
|
|
| 62 | c2l37A_ |
|
not modelled |
5.9 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribosome-inactivating protein luffin p1;
PDBTitle: 3d solution structure of arginine/glutamate-rich polypeptide luffin p12 from the seeds of sponge gourd (luffa cylindrical)
|
|
|
| 63 | c3r66A_ |
|
not modelled |
5.7 |
23 |
PDB header:viral protein/antiviral protein
Chain: A: PDB Molecule:non-structural protein 1;
PDBTitle: crystal structure of human isg15 in complex with ns1 n-terminal region2 from influenza virus b, northeast structural genomics consortium3 target ids hx6481, hr2873, and or2
|
|
|
| 64 | d2hxja1 |
|
not modelled |
5.7 |
18 |
Fold:FinO-like
Superfamily:FinO-like
Family:FinO-like |
|
|
| 65 | c6fxdB_ |
|
not modelled |
5.7 |
15 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:mupz;
PDBTitle: crystal structure of mupz from pseudomonas fluorescens
|
|
|
| 66 | d1iq4a_ |
|
not modelled |
5.6 |
20 |
Fold:RL5-like
Superfamily:RL5-like
Family:Ribosomal protein L5 |
|
|
| 67 | d2zjrd1 |
|
not modelled |
5.5 |
18 |
Fold:RL5-like
Superfamily:RL5-like
Family:Ribosomal protein L5 |
|
|
| 68 | c6o3sA_ |
|
not modelled |
5.5 |
36 |
PDB header:plant protein, hydrolase
Chain: A: PDB Molecule:ribosome-inactivating protein luffin p1;
PDBTitle: nmr solution structure of vicilin-buried peptide-8 (vbp-8)
|
|
|
| 69 | d1ei7a_ |
|
not modelled |
5.3 |
10 |
Fold:Four-helical up-and-down bundle
Superfamily:TMV-like viral coat proteins
Family:TMV-like viral coat proteins |
|
|
| 70 | d1v3fa_ |
|
not modelled |
5.2 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:DEP domain |
|
|
| 71 | c2hxjF_ |
|
not modelled |
5.2 |
18 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a protein of unknown function nmb1681 from2 neisseria meningitidis mc58, possible nucleic acid binding protein
|
|
|
| 72 | d1a7ia1 |
|
not modelled |
5.2 |
10 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
|
|
| 73 | c3htrB_ |
|
not modelled |
5.1 |
32 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized prc-barrel domain protein;
PDBTitle: crystal structure of prc-barrel domain protein from2 rhodopseudomonas palustris
|
|
|