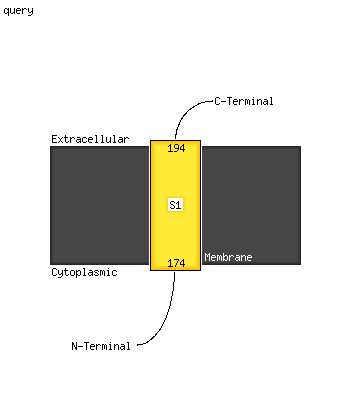
| 1 | c3pe5B_
|
|
 |
100.0 |
27 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: three-dimensional structure of protein a7vv38_9clot from clostridium2 leptum dsm 753, northeast structural genomics consortium target3 qlr103
|
|
|
|
| 2 | c5v8cA_
|
|
 |
100.0 |
25 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator;
PDBTitle: lytr-csp2a-psr enzyme from actinomyces oris
|
|
|
|
| 3 | c3okzB_
|
|
 |
100.0 |
22 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative uncharacterized protein gbs0355;
PDBTitle: crystal structure of protein gbs0355 from streptococcus agalactiae,2 northeast structural genomics consortium target sar127
|
|
|
|
| 4 | c4de8A_
|
|
 |
100.0 |
23 |
PDB header:membrane protein
Chain: A: PDB Molecule:cps2a;
PDBTitle: lytr-cps2a-psr family protein with bound octaprenyl monophosphate2 lipid
|
|
|
|
| 5 | c3qfiA_
|
|
 |
100.0 |
24 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator;
PDBTitle: x-ray crystal structure of transcriptional regulator (ef0465) from2 enterococcus faecalis, northeast structural genomics consortium3 target efr190
|
|
|
|
| 6 | c3owqB_
|
|
 |
100.0 |
30 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:lin1025 protein;
PDBTitle: x-ray structure of lin1025 protein from listeria innocua, northeast2 structural genomics consortium target lkr164
|
|
|
|
| 7 | c3mejA_
|
|
 |
100.0 |
26 |
PDB header:transcriptional regulator
Chain: A: PDB Molecule:transcriptional regulator ywtf;
PDBTitle: crystal structure of putative transcriptional regulator ywtf from2 bacillus subtilis, northeast structural genomics consortium target3 sr736
|
|
|
|
| 8 | c3nroA_
|
|
 |
100.0 |
30 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lmo1026 protein;
PDBTitle: crystal structure of putative transcriptional factor lmo1026 from2 listeria monocytogenes (fragment 52-321), northeast structural3 genomics consortium target lmr194
|
|
|
|
| 9 | c4obmA_
|
|
 |
100.0 |
14 |
PDB header:transcription regulator
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative transcription regulator (eubsir_01389)2 from eubacterium siraeum dsm 15702 at 2.15 a resolution
|
|
|
|
| 10 | c3nxhA_
|
|
 |
100.0 |
22 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator yvhj;
PDBTitle: crystal structure of the transcriptional regulator yvhj from bacillus2 subtilis. northeast structural genomics consortium target sr735.
|
|
|
|
| 11 | c2m5yA_
|
|
 |
99.7 |
22 |
PDB header:unknown function
Chain: A: PDB Molecule:putative tuberculin related peptide;
PDBTitle: solution structure of the c-terminal domain of rv0431
|
|
|
|
| 12 | c2pjuD_
|
|
 |
87.2 |
15 |
PDB header:transcription
Chain: D: PDB Molecule:propionate catabolism operon regulatory protein;
PDBTitle: crystal structure of propionate catabolism operon regulatory protein2 prpr
|
|
|
|
| 13 | c3txsC_
|
|
 |
67.5 |
18 |
PDB header:viral protein
Chain: C: PDB Molecule:terminase dna packaging enzyme small subunit;
PDBTitle: crystal structure of phage 44rr small terminase gp16
|
|
|
|
| 14 | d1y81a1
|
|
 |
24.0 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
|
| 15 | c1oheA_
|
|
 |
22.4 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:cdc14b2 phosphatase;
PDBTitle: structure of cdc14b phosphatase with a peptide ligand
|
|
|
|
| 16 | c5j5vC_
|
|
 |
18.1 |
29 |
PDB header:toxin
Chain: C: PDB Molecule:immunity protein cdii;
PDBTitle: cdia-ct from uropathogenic escherichia coli in complex with cognate2 immunity protein and cysk
|
|
|
|
| 17 | c4ddpA_
|
|
 |
17.3 |
18 |
PDB header:membrane protein
Chain: A: PDB Molecule:beclin-1;
PDBTitle: crystal structure of beclin 1 evolutionarily conserved domain(ecd)
|
|
|
|
| 18 | d1ny8a_
|
|
 |
16.7 |
24 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:BolA-like
Family:BolA-like |
|
|
|
| 19 | c6f0kA_
|
|
 |
16.5 |
9 |
PDB header:membrane protein
Chain: A: PDB Molecule:cytochrome c family protein;
PDBTitle: alternative complex iii
|
|
|
|
| 20 | d1gsoa2
|
|
 |
16.2 |
25 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
|
|
|
| 21 | d1xjca_ |
|
not modelled |
16.1 |
20 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
| 22 | c2ppwA_ |
|
not modelled |
16.0 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:conserved domain protein;
PDBTitle: the crystal structure of uncharacterized ribose 5-phosphate isomerase2 rpib from streptococcus pneumoniae
|
|
|
| 23 | d1vkza2 |
|
not modelled |
14.3 |
17 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
|
|
| 24 | d1ohea2 |
|
not modelled |
13.4 |
20 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Dual specificity phosphatase-like |
|
|
| 25 | c5iiqA_ |
|
not modelled |
12.7 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:vacuolar transporter chaperone 4;
PDBTitle: structure of the spx-ttm domain fragment of the yeast inorganic2 polyphophate polymerase vtc4 (form b).
|
|
|
| 26 | c5z5bA_ |
|
not modelled |
12.7 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein-tyrosine phosphatase;
PDBTitle: crystal structure of tk-ptp in the g95a mutant form
|
|
|
| 27 | c3bnuA_ |
|
not modelled |
11.9 |
32 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:polyamine oxidase fms1;
PDBTitle: crystal structure of polyamine oxidase fms1 from2 saccharomyces cerevisiae in complex with bis-(3s,3's)-3 methylated spermine
|
|
|
| 28 | c4qn9A_ |
|
not modelled |
11.7 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-acyl-phosphatidylethanolamine-hydrolyzing phospholipase
PDBTitle: structure of human nape-pld
|
|
|
| 29 | c3vp7A_ |
|
not modelled |
11.4 |
18 |
PDB header:protein transport
Chain: A: PDB Molecule:vacuolar protein sorting-associated protein 30;
PDBTitle: crystal structure of the beta-alpha repeated, autophagy-specific2 (bara) domain of vps30/atg6
|
|
|
| 30 | d1tf4a2 |
|
not modelled |
11.3 |
30 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:Carbohydrate-binding domain
Family:Cellulose-binding domain family III |
|
|
| 31 | d2cdqa3 |
|
not modelled |
11.2 |
16 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
|
|
| 32 | c4b96A_ |
|
not modelled |
11.1 |
10 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:cellulose binding domain-containing protein;
PDBTitle: family 3b carbohydrate-binding module from the biomass2 sensoring system of clostridium clariflavum
|
|
|
| 33 | c2duwA_ |
|
not modelled |
10.8 |
14 |
PDB header:ligand binding protein
Chain: A: PDB Molecule:putative coa-binding protein;
PDBTitle: solution structure of putative coa-binding protein of2 klebsiella pneumoniae
|
|
|
| 34 | c4ecgA_ |
|
not modelled |
10.7 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative iron-regulated protein a;
PDBTitle: crystal structure of a putative iron-regulated protein a precursor2 (bdi_2603) from parabacteroides distasonis atcc 8503 at 2.30 a3 resolution
|
|
|
| 35 | c4alnE_ |
|
not modelled |
10.7 |
13 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:enoyl-[acyl-carrier-protein] reductase [nadph];
PDBTitle: crystal structure of s. aureus fabi (p32)
|
|
|
| 36 | d2d59a1 |
|
not modelled |
10.0 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
| 37 | c4puiA_ |
|
not modelled |
9.8 |
24 |
PDB header:protein binding
Chain: A: PDB Molecule:sufe-like protein, chloroplastic;
PDBTitle: bola domain of sufe1 from arabidopsis thaliana
|
|
|
| 38 | c3k31B_ |
|
not modelled |
9.7 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:enoyl-(acyl-carrier-protein) reductase;
PDBTitle: crystal structure of eonyl-(acyl-carrier-protein) reductase from2 anaplasma phagocytophilum in complex with nad at 1.9a resolution
|
|
|
| 39 | c4nk4E_ |
|
not modelled |
9.7 |
9 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:enoyl-[acyl-carrier-protein] reductase [nadh];
PDBTitle: crystal structure of fabi from candidatus liberibacter asiaticus
|
|
|
| 40 | c4b9cA_ |
|
not modelled |
9.5 |
30 |
PDB header:carbohydrate-binding protein
Chain: A: PDB Molecule:type 3a cellulose-binding domain protein;
PDBTitle: biomass sensoring modules from putative rsgi-like proteins2 of clostridium thermocellum resemble family 3 carbohydrate-3 binding module of cellulosome
|
|
|
| 41 | c3ff4A_ |
|
not modelled |
9.5 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein chu_1412
|
|
|
| 42 | d1nbca_ |
|
not modelled |
9.4 |
30 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:Carbohydrate-binding domain
Family:Cellulose-binding domain family III |
|
|
| 43 | c3o2eA_ |
|
not modelled |
9.4 |
29 |
PDB header:unknown function
Chain: A: PDB Molecule:bola-like protein;
PDBTitle: crystal structure of a bol-like protein from babesia bovis
|
|
|
| 44 | c3grkE_ |
|
not modelled |
9.4 |
6 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:enoyl-(acyl-carrier-protein) reductase (nadh);
PDBTitle: crystal structure of short chain dehydrogenase reductase2 sdr glucose-ribitol dehydrogenase from brucella melitensis
|
|
|
| 45 | c2fgyA_ |
|
not modelled |
9.2 |
39 |
PDB header:lyase
Chain: A: PDB Molecule:carboxysome shell polypeptide;
PDBTitle: beta carbonic anhydrase from the carboxysomal shell of2 halothiobacillus neapolitanus (csosca)
|
|
|
| 46 | c2dhmA_ |
|
not modelled |
9.2 |
14 |
PDB header:protein binding
Chain: A: PDB Molecule:protein bola;
PDBTitle: solution structure of the bola protein from escherichia coli
|
|
|
| 47 | c2xfgB_ |
|
not modelled |
9.1 |
20 |
PDB header:hydrolase/sugar binding protein
Chain: B: PDB Molecule:endoglucanase 1;
PDBTitle: reassembly and co-crystallization of a family 9 processive2 endoglucanase from separately expressed gh9 and cbm3c3 modules
|
|
|
| 48 | c1u8sB_ |
|
not modelled |
8.9 |
13 |
PDB header:transcription
Chain: B: PDB Molecule:glycine cleavage system transcriptional
PDBTitle: crystal structure of putative glycine cleavage system2 transcriptional repressor
|
|
|
| 49 | d2hmfa2 |
|
not modelled |
8.8 |
15 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
|
|
| 50 | c5ijpA_ |
|
not modelled |
8.8 |
13 |
PDB header:inositol phosphate binding protein
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the spx domain of chaetomium thermophilum vtc4 in2 complex with inositol hexakisphosphate (insp6).
|
|
|
| 51 | d1t70a_ |
|
not modelled |
8.8 |
21 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:DR1281-like |
|
|
| 52 | c5iitC_ |
|
not modelled |
8.8 |
13 |
PDB header:inositol phosphate binding protein
Chain: C: PDB Molecule:vacuolar transporter chaperone 4,core histone macro-h2a.1;
PDBTitle: structure of spx domain of the yeast inorganic polyphophate polymerase2 vtc4 crystallized by carrier-driven crystallization in fusion with3 the macro domain of human histone macroh2a1.1
|
|
|
| 53 | c1wv9B_ |
|
not modelled |
8.7 |
18 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:rhodanese homolog tt1651;
PDBTitle: crystal structure of rhodanese homolog tt1651 from an2 extremely thermophilic bacterium thermus thermophilus hb8
|
|
|
| 54 | c5ijjB_ |
|
not modelled |
8.7 |
24 |
PDB header:inositol polyphosphate binding protein
Chain: B: PDB Molecule:spx domain;
PDBTitle: structure of the spx domain of chaetomium thermophilum2 glycerophosphodiester phosphodiesterase 1 in complex with inositol3 hexakisphosphate (insp6)
|
|
|
| 55 | d2z06a1 |
|
not modelled |
8.7 |
18 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:TTHA0625-like |
|
|
| 56 | c3i7aA_ |
|
not modelled |
8.7 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative metal-dependent phosphohydrolase;
PDBTitle: crystal structure of putative metal-dependent phosphohydrolase2 (yp_926882.1) from shewanella amazonensis sb2b at 2.06 a resolution
|
|
|
| 57 | c6d5bL_ |
|
not modelled |
8.4 |
20 |
PDB header:sugar binding protein
Chain: L: PDB Molecule:glycoside hydrolase wp_045175321;
PDBTitle: structure of caldicellulosiruptor danielii cbm3 module of glycoside2 hydrolase wp_045175321
|
|
|
| 58 | d1iaza_ |
|
not modelled |
8.4 |
22 |
Fold:Cytolysin/lectin
Superfamily:Cytolysin/lectin
Family:Anemone pore-forming cytolysin |
|
|
| 59 | c2nclA_ |
|
not modelled |
8.4 |
10 |
PDB header:protein binding
Chain: A: PDB Molecule:bola-like protein 3;
PDBTitle: solution structure of bola3 from homo sapiens
|
|
|
| 60 | c2l8aA_ |
|
not modelled |
8.4 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:endoglucanase;
PDBTitle: structure of a novel cbm3 lacking the calcium-binding site
|
|
|
| 61 | c4pugA_ |
|
not modelled |
8.4 |
19 |
PDB header:dna binding protein
Chain: A: PDB Molecule:bola like protein;
PDBTitle: bola1 from arabidopsis thaliana
|
|
|
| 62 | d1g87a2 |
|
not modelled |
8.3 |
20 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:Carbohydrate-binding domain
Family:Cellulose-binding domain family III |
|
|
| 63 | c2o8sA_ |
|
not modelled |
8.3 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:agr_c_984p;
PDBTitle: x-ray crystal structure of protein agr_c_984 from agrobacterium2 tumefaciens. northeast structural genomics consortium target atr120.
|
|
|
| 64 | c2jb1B_ |
|
not modelled |
8.1 |
35 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-amino acid oxidase;
PDBTitle: the l-amino acid oxidase from rhodococcus opacus in complex2 with l-alanine
|
|
|
| 65 | c2xbtA_ |
|
not modelled |
8.0 |
30 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:cellulosomal scaffoldin;
PDBTitle: structure of a scaffoldin carbohydrate-binding module family 3b from2 the cellulosome of bacteroides cellulosolvens: structural diversity3 and implications for carbohydrate binding
|
|
|
| 66 | d1vqon1 |
|
not modelled |
8.0 |
16 |
Fold:Ribonuclease H-like motif
Superfamily:Translational machinery components
Family:Ribosomal protein L18 and S11 |
|
|
| 67 | c4q9nD_ |
|
not modelled |
7.9 |
9 |
PDB header:oxidoreductase/oxidoreductase inhibitor
Chain: D: PDB Molecule:enoyl-[acyl-carrier-protein] reductase [nadh];
PDBTitle: crystal structure of chlamydia trachomatis enoyl-acp reductase (fabi)2 in complex with nadh and afn-1252
|
|
|
| 68 | d1fnna1 |
|
not modelled |
7.9 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Helicase DNA-binding domain |
|
|
| 69 | c2ozeA_ |
|
not modelled |
7.9 |
15 |
PDB header:dna binding protein
Chain: A: PDB Molecule:orf delta';
PDBTitle: the crystal structure of delta protein of psm19035 from2 streptoccocus pyogenes
|
|
|
| 70 | c3l76B_ |
|
not modelled |
7.9 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:aspartokinase;
PDBTitle: crystal structure of aspartate kinase from synechocystis
|
|
|
| 71 | c2e1mA_ |
|
not modelled |
7.7 |
36 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-glutamate oxidase;
PDBTitle: crystal structure of l-glutamate oxidase from streptomyces sp. x-119-6
|
|
|
| 72 | c2n5iA_ |
|
not modelled |
7.6 |
19 |
PDB header:transport protein
Chain: A: PDB Molecule:peptidyl carrier protein pltl;
PDBTitle: pltl-pyrrolyl
|
|
|
| 73 | d1v86a_ |
|
not modelled |
7.4 |
31 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
|
|
| 74 | c5xw4A_ |
|
not modelled |
7.3 |
11 |
PDB header:cell cycle
Chain: A: PDB Molecule:tyrosine-protein phosphatase cdc14;
PDBTitle: crystal structure of budding yeast cdc14p (wild type) in the apo state
|
|
|
| 75 | c5nfmA_ |
|
not modelled |
7.3 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:yrba;
PDBTitle: crystal structure of yrba from sinorhizobium meliloti in complex with2 copper.
|
|
|
| 76 | c2qnwA_ |
|
not modelled |
7.2 |
19 |
PDB header:signaling protein
Chain: A: PDB Molecule:acyl carrier protein;
PDBTitle: toxoplasma gondii apicoplast-targeted acyl carrier protein
|
|
|
| 77 | d1iuka_ |
|
not modelled |
7.0 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
| 78 | c2ylkD_ |
|
not modelled |
6.9 |
10 |
PDB header:hydrolase
Chain: D: PDB Molecule:cellulose 1,4-beta-cellobiosidase;
PDBTitle: carbohydrate-binding module cbm3b from the cellulosomal2 cellobiohydrolase 9a from clostridium thermocellum
|
|
|
| 79 | c2yg4B_ |
|
not modelled |
6.9 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putrescine oxidase;
PDBTitle: structure-based redesign of cofactor binding in putrescine2 oxidase: wild type bound to putrescine
|
|
|
| 80 | c3gdfA_ |
|
not modelled |
6.9 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable nadp-dependent mannitol dehydrogenase;
PDBTitle: crystal structure of the nadp-dependent mannitol dehydrogenase from2 cladosporium herbarum.
|
|
|
| 81 | c1b22A_ |
|
not modelled |
6.8 |
31 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna repair protein rad51;
PDBTitle: rad51 (n-terminal domain)
|
|
|
| 82 | d1b22a_ |
|
not modelled |
6.8 |
31 |
Fold:SAM domain-like
Superfamily:Rad51 N-terminal domain-like
Family:DNA repair protein Rad51, N-terminal domain |
|
|
| 83 | d1f80d_ |
|
not modelled |
6.8 |
16 |
Fold:Acyl carrier protein-like
Superfamily:ACP-like
Family:Acyl-carrier protein (ACP) |
|
|
| 84 | d1e6ya2 |
|
not modelled |
6.7 |
23 |
Fold:Ferredoxin-like
Superfamily:Methyl-coenzyme M reductase subunits
Family:Methyl-coenzyme M reductase alpha and beta chain N-terminal domain |
|
|
| 85 | c3ce7A_ |
|
not modelled |
6.6 |
10 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:specific mitochondrial acyl carrier protein;
PDBTitle: crystal structure of toxoplasma specific mitochondrial acyl carrier2 protein, 59.m03510
|
|
|
| 86 | c2mm9A_ |
|
not modelled |
6.6 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:bola2;
PDBTitle: solution structure of reduced bola2 from arabidopsis thaliana
|
|
|
| 87 | c2f06B_ |
|
not modelled |
6.6 |
10 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:conserved hypothetical protein;
PDBTitle: crystal structure of protein bt0572 from bacteroides thetaiotaomicron
|
|
|
| 88 | c4m87B_ |
|
not modelled |
6.6 |
6 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:enoyl-[acyl-carrier-protein] reductase [nadh];
PDBTitle: crystal structure of enoyl-acyl carrier protein reductase (fabi) from2 neisseria meningitidis in complex with nad+
|
|
|
| 89 | c1n7sB_ |
|
not modelled |
6.5 |
17 |
PDB header:transport protein
Chain: B: PDB Molecule:syntaxin 1a;
PDBTitle: high resolution structure of a truncated neuronal snare complex
|
|
|
| 90 | c3un1D_ |
|
not modelled |
6.5 |
12 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:probable oxidoreductase;
PDBTitle: crystal structure of an oxidoreductase from sinorhizobium meliloti2 1021
|
|
|
| 91 | d2voua1 |
|
not modelled |
6.5 |
24 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 92 | c2zuuA_ |
|
not modelled |
6.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:lacto-n-biose phosphorylase;
PDBTitle: crystal structure of galacto-n-biose/lacto-n-biose i phosphorylase in2 complex with glcnac
|
|
|
| 93 | c4iyrB_ |
|
not modelled |
6.4 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:caspase-6;
PDBTitle: crystal structure of full-length caspase-6 zymogen
|
|
|
| 94 | c2lfvA_ |
|
not modelled |
6.4 |
8 |
PDB header:cell cycle
Chain: A: PDB Molecule:protein damx;
PDBTitle: solution structure of the spor domain from e. coli damx
|
|
|
| 95 | c1v60A_ |
|
not modelled |
6.4 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:riken cdna 1810037g04;
PDBTitle: solution structure of bola1 protein from mus musculus
|
|
|
| 96 | c5ldgA_ |
|
not modelled |
6.4 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:(-)-isopiperitenone reductase;
PDBTitle: isopiperitenone reductase from mentha piperita in complex with2 isopiperitenone and nadp
|
|
|
| 97 | c3gr6A_ |
|
not modelled |
6.3 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl-[acyl-carrier-protein] reductase [nadh];
PDBTitle: crystal structure of the staphylococcus aureus enoyl-acyl carrier2 protein reductase (fabi) in complex with nadp and triclosan
|
|
|
| 98 | c5y4bA_ |
|
not modelled |
6.3 |
5 |
PDB header:electron transport
Chain: A: PDB Molecule:bola-like protein 2;
PDBTitle: solution structure of yeast fra2
|
|
|
| 99 | c4ne4A_ |
|
not modelled |
6.3 |
19 |
PDB header:transport protein
Chain: A: PDB Molecule:abc transporter, substrate binding protein
PDBTitle: crystal structure of abc transporter substrate binding protein prox2 from agrobacterium tumefaciens cocrystalized with btb
|
|
|