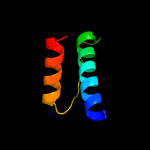
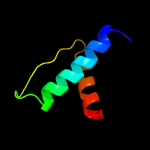
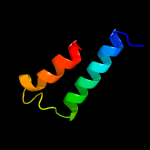
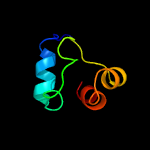
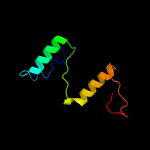
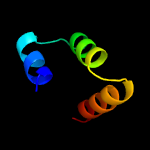
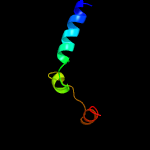
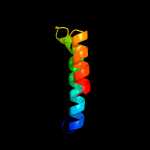
1 c5o1mA_
99.3
15
PDB header: oxidoreductaseChain: A: PDB Molecule: rubber oxygenase;PDBTitle: structure of latex clearing protein lcp in the closed state
2 c2ru8A_
39.1
13
PDB header: replicationChain: A: PDB Molecule: primosomal protein 1;PDBTitle: dnat c-terminal domain
3 c4ou6A_
27.9
13
PDB header: replication/dnaChain: A: PDB Molecule: primosomal protein 1;PDBTitle: crystal structure of dnat84-153-dt10 ssdna complex form 1
4 c5cuvB_
20.4
20
PDB header: metal binding proteinChain: B: PDB Molecule: acidocalcisomal pyrophosphatase;PDBTitle: crystal structure of trypanosoma cruzi vacuolar soluble2 pyrophosphatases in apo form
5 d1g73a_
16.2
4
Fold: Spectrin repeat-likeSuperfamily: Smac/diabloFamily: Smac/diablo
6 c4plaA_
14.8
27
PDB header: transferase,hydrolaseChain: A: PDB Molecule: chimera protein of phosphatidylinositol 4-kinase type 2-PDBTitle: crystal structure of phosphatidyl inositol 4-kinase ii alpha in2 complex with atp
7 c1unhD_
14.0
6
PDB header: cell cycleChain: D: PDB Molecule: cyclin-dependent kinase 5 activator 1;PDBTitle: structural mechanism for the inhibition of cdk5-p25 by2 roscovitine, aloisine and indirubin.
8 d1unld_
13.6
6
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Cyclin
9 c4nk1B_
13.0
16
PDB header: oxygen bindingChain: B: PDB Molecule: hemoglobin-like protein;PDBTitle: crystal structure of phosphate-bound hell's gate globin iv
10 c2e8mA_
12.3
22
PDB header: signaling proteinChain: A: PDB Molecule: epidermal growth factor receptor kinasePDBTitle: solution structure of the c-terminal sam-domain of2 epidermal growth receptor pathway substrate 8
11 c4wiuA_
11.0
18
PDB header: lyase,transferaseChain: A: PDB Molecule: phosphoenolpyruvate carboxykinase [gtp];PDBTitle: crystal structure of pepck (rv0211) from mycobacterium tuberculosis in2 complex with oxalate and mn2+
12 d1fewa_
10.5
4
Fold: Spectrin repeat-likeSuperfamily: Smac/diabloFamily: Smac/diablo
13 d1u7ka_
9.9
10
Fold: Retrovirus capsid protein, N-terminal core domainSuperfamily: Retrovirus capsid protein, N-terminal core domainFamily: Retrovirus capsid protein, N-terminal core domain
14 c6fviA_
8.5
46
PDB header: cell cycleChain: A: PDB Molecule: centrosomal protein of 192 kda;PDBTitle: ash / papd-like domain of human cep192 (papd-like domain 7)
15 c2jspA_
8.2
14
PDB header: gene regulationChain: A: PDB Molecule: transcriptional regulatory protein ros;PDBTitle: the prokaryotic cys2his2 zinc finger adopts a novel fold as2 revealed by the nmr structure of a. tumefaciens ros dna3 binding domain
16 c1nexC_
7.8
12
PDB header: ligase, cell cycleChain: C: PDB Molecule: centromere dna-binding protein complex cbf3PDBTitle: crystal structure of scskp1-sccdc4-cpd peptide complex
17 d1v7ba2
7.8
9
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain
18 c1mk2B_
7.7
44
PDB header: transcriptionChain: B: PDB Molecule: madh-interacting protein;PDBTitle: smad3 sbd complex
19 c4do7B_
7.6
23
PDB header: hydrolaseChain: B: PDB Molecule: amidohydrolase 2;PDBTitle: crystal structure of an amidohydrolase (cog3618) from burkholderia2 multivorans (target efi-500235) with bound zn, space group c2
20 c6alyA_
7.4
13
PDB header: transcriptionChain: A: PDB Molecule: mediator of rna polymerase ii transcription subunit 15;PDBTitle: solution structure of yeast med15 abd2 residues 277-368
21 c3m20A_
not modelled
7.3
17
PDB header: isomeraseChain: A: PDB Molecule: 4-oxalocrotonate tautomerase, putative;PDBTitle: crystal structure of dmpi from archaeoglobus fulgidus determined to2 2.37 angstroms resolution
22 c3ucsB_
not modelled
7.3
8
PDB header: chaperoneChain: B: PDB Molecule: chaperone-modulator protein cbpm;PDBTitle: crystal structure of the complex between cbpa j-domain and cbpm
23 c4umoB_
not modelled
7.1
26
PDB header: signaling proteinChain: B: PDB Molecule: potassium voltage-gated channel subfamily kqt member 1;PDBTitle: crystal structure of the kv7.1 proximal c-terminal domain in complex2 with calmodulin
24 d16vpa_
not modelled
7.0
13
Fold: Conserved core of transcriptional regulatory protein vp16Superfamily: Conserved core of transcriptional regulatory protein vp16Family: Conserved core of transcriptional regulatory protein vp16
25 d1xg7a_
not modelled
6.6
56
Fold: DNA-glycosylaseSuperfamily: DNA-glycosylaseFamily: AgoG-like
26 c3a08C_
not modelled
6.5
41
PDB header: structural proteinChain: C: PDB Molecule: collagen-like peptide;PDBTitle: structure of (ppg)4-oog-(ppg)4, monoclinic, twinned crystal
27 c2glwA_
not modelled
6.4
15
PDB header: transcriptionChain: A: PDB Molecule: 92aa long hypothetical protein;PDBTitle: the solution structure of phs018 from pyrococcus horikoshii
28 c2ovqA_
not modelled
6.3
14
PDB header: transcription/cell cycleChain: A: PDB Molecule: s-phase kinase-associated protein 1a;PDBTitle: structure of the skp1-fbw7-cyclinedegc complex
29 d1iwma_
not modelled
6.1
33
Fold: LolA-like prokaryotic lipoproteins and lipoprotein localization factorsSuperfamily: Prokaryotic lipoproteins and lipoprotein localization factorsFamily: Outer membrane lipoprotein receptor LolB
30 c4fdxB_
not modelled
5.9
19
PDB header: isomeraseChain: B: PDB Molecule: 4-oxalocrotonase tautomerase isozyme;PDBTitle: kinetic and structural characterization of the 4-oxalocrotonate2 tautomerase isozymes from methylibium petroleiphilum
31 c3a08F_
not modelled
5.9
50
PDB header: structural proteinChain: F: PDB Molecule: collagen-like peptide;PDBTitle: structure of (ppg)4-oog-(ppg)4, monoclinic, twinned crystal
32 c4okmA_
not modelled
5.8
19
PDB header: transferaseChain: A: PDB Molecule: terpene synthase metal-binding domain-containing protein;PDBTitle: selinadiene synthase apo and in complex with diphosphate
33 c2bmmA_
not modelled
5.7
9
PDB header: oxygen storage/transportChain: A: PDB Molecule: thermostable hemoglobin from thermobifida fusca;PDBTitle: x-ray structure of a novel thermostable hemoglobin from the2 actinobacterium thermobifida fusca
34 c3zs9D_
not modelled
5.6
53
PDB header: hydrolase/transport proteinChain: D: PDB Molecule: golgi to er traffic protein 2;PDBTitle: s. cerevisiae get3-adp-alf4- complex with a cytosolic get2 fragment
35 d2gica1
not modelled
5.5
26
Fold: Rhabdovirus nucleoprotein-likeSuperfamily: Rhabdovirus nucleoprotein-likeFamily: Rhabdovirus nucleocapsid protein
36 c5k21C_
not modelled
5.5
71
PDB header: oxidoreductaseChain: C: PDB Molecule: pyocyanin demethylase;PDBTitle: pyocyanin demethylase
37 d1q3ma_
not modelled
5.5
31
Fold: GLA-domainSuperfamily: GLA-domainFamily: GLA-domain
38 d2g3ba2
not modelled
5.4
9
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain
39 c3a08E_
not modelled
5.3
50
PDB header: structural proteinChain: E: PDB Molecule: collagen-like peptide;PDBTitle: structure of (ppg)4-oog-(ppg)4, monoclinic, twinned crystal
40 c2d3hF_
not modelled
5.3
50
PDB header: structural proteinChain: F: PDB Molecule: collagen model peptides (pro-pro-gly)4-hyp-hyp-PDBTitle: crystal structures of collagen model peptides (pro-pro-gly)2 4-hyp-hyp-gly-(pro-pro-gly)4
41 c5yo8B_
not modelled
5.3
22
PDB header: lyaseChain: B: PDB Molecule: tetraprenyl-beta-curcumene synthase;PDBTitle: crystal structure of beta-c25/c30/c35-prene synthase
42 c2zc2A_
not modelled
5.2
17
PDB header: replicationChain: A: PDB Molecule: dnad-like replication protein;PDBTitle: crystal structure of dnad-like replication protein from streptococcus2 mutans ua159, gi 24377835, residues 127-199
43 d1eyxa_
not modelled
5.1
13
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins
44 c4ue8B_
not modelled
5.1
39
PDB header: translationChain: B: PDB Molecule: 4e-binding protein thor;PDBTitle: complex of d. melanogaster eif4e with the 4e binding protein thor
45 d1nb4a_
not modelled
5.1
13
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: RNA-dependent RNA-polymerase
46 c2d3hC_
not modelled
5.1
43
PDB header: structural proteinChain: C: PDB Molecule: collagen model peptides (pro-pro-gly)4-hyp-hyp-PDBTitle: crystal structures of collagen model peptides (pro-pro-gly)2 4-hyp-hyp-gly-(pro-pro-gly)4