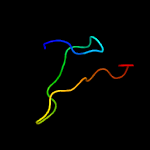
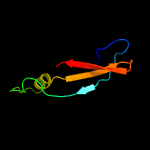
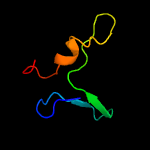
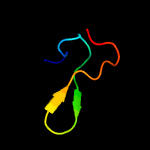
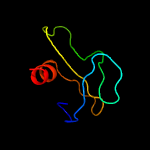
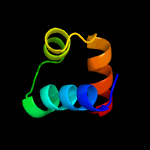
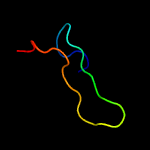
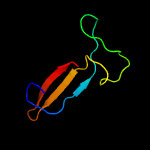
1 c3gn5B_
93.7
13
PDB header: dna binding proteinChain: B: PDB Molecule: hth-type transcriptional regulator mqsa (ygit/b3021);PDBTitle: structure of the e. coli protein mqsa (ygit/b3021)
2 c2qkdA_
89.1
19
PDB header: signaling protein, cell cycleChain: A: PDB Molecule: zinc finger protein zpr1;PDBTitle: crystal structure of tandem zpr1 domains
3 c2hu9B_
87.6
19
PDB header: metal transportChain: B: PDB Molecule: mercuric transport protein periplasmic component;PDBTitle: x-ray structure of the archaeoglobus fulgidus copz n-2 terminal domain
4 d1dgsa1
87.1
32
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: NAD+-dependent DNA ligase, domain 3
5 c5fjaI_
78.5
32
PDB header: transcriptionChain: I: PDB Molecule: dna-directed rna polymerase iii subunit rpc10;PDBTitle: cryo-em structure of yeast rna polymerase iii at 4.7 a
6 d1pvma3
76.7
39
Fold: Rubredoxin-likeSuperfamily: Hypothetical protein Ta0289 C-terminal domainFamily: Hypothetical protein Ta0289 C-terminal domain
7 c1dgsB_
74.7
32
PDB header: ligaseChain: B: PDB Molecule: dna ligase;PDBTitle: crystal structure of nad+-dependent dna ligase from t.2 filiformis
8 d2fiya1
73.3
29
Fold: FdhE-likeSuperfamily: FdhE-likeFamily: FdhE-like
9 c1v9pB_
69.3
32
PDB header: ligaseChain: B: PDB Molecule: dna ligase;PDBTitle: crystal structure of nad+-dependent dna ligase
10 c5flmI_
65.7
24
PDB header: transcriptionChain: I: PDB Molecule: dna-directed rna polymerase ii subunit rpb9;PDBTitle: structure of transcribing mammalian rna polymerase ii
11 d1x3za1
64.0
16
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core
12 c3floD_
59.5
18
PDB header: transferaseChain: D: PDB Molecule: dna polymerase alpha catalytic subunit a;PDBTitle: crystal structure of the carboxyl-terminal domain of yeast dna2 polymerase alpha in complex with its b subunit
13 d1k78a1
58.0
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain
14 c2owoA_
57.1
32
PDB header: ligase/dnaChain: A: PDB Molecule: dna ligase;PDBTitle: last stop on the road to repair: structure of e.coli dna ligase bound2 to nicked dna-adenylate
15 c4qiwP_
54.0
28
PDB header: transcriptionChain: P: PDB Molecule: dna-directed rna polymerase subunit p;PDBTitle: crystal structure of euryarchaeal rna polymerase from thermococcus2 kodakarensis
16 c4glxA_
53.0
37
PDB header: ligase/ligase inhibitor/dnaChain: A: PDB Molecule: dna ligase;PDBTitle: dna ligase a in complex with inhibitor
17 d1qpza1
52.0
17
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator
18 d1qypa_
51.0
29
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain
19 d2dk5a1
50.9
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RPO3F domain-like
20 d1twfi2
50.5
13
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain
21 c3j39p_
not modelled
49.9
23
PDB header: ribosomeChain: P: PDB Molecule: 60s ribosomal protein l17;PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
22 d2hsga1
not modelled
47.7
8
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator
23 d1lcda_
not modelled
47.5
4
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator
24 d2bjca1
not modelled
46.7
4
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator
25 d1uxca_
not modelled
45.7
9
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator
26 d1hlva1
not modelled
44.9
29
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Centromere-binding
27 d1a1ha1
not modelled
44.3
33
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
28 c2lcvA_
not modelled
44.2
18
PDB header: transcription regulatorChain: A: PDB Molecule: hth-type transcriptional repressor cytr;PDBTitle: structure of the cytidine repressor dna-binding domain; an alternate2 calculation
29 c3izrm_
not modelled
43.4
33
PDB header: ribosomeChain: M: PDB Molecule: 60s ribosomal protein l23 (l14p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
30 d1efaa1
not modelled
43.3
4
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator
31 d2jnea1
not modelled
43.0
33
Fold: Rubredoxin-likeSuperfamily: YfgJ-likeFamily: YfgJ-like
32 c2jneA_
not modelled
43.0
33
PDB header: metal binding proteinChain: A: PDB Molecule: hypothetical protein yfgj;PDBTitle: nmr structure of e.coli yfgj modelled with two zn+2 bound. northeast2 structural genomics consortium target er317.
33 d1uxda_
not modelled
43.0
9
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: GalR/LacI-like bacterial regulator
34 c4bs9A_
not modelled
42.6
36
PDB header: isomeraseChain: A: PDB Molecule: trud;PDBTitle: structure of the heterocyclase trud
35 d2akla2
not modelled
42.2
43
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: PhnA zinc-binding domain
36 c3h0gI_
not modelled
41.5
24
PDB header: transcriptionChain: I: PDB Molecule: dna-directed rna polymerase ii subunit rpb9;PDBTitle: rna polymerase ii from schizosaccharomyces pombe
37 c5a3aA_
not modelled
41.5
36
PDB header: transferaseChain: A: PDB Molecule: sir2 family protein;PDBTitle: crystal structure of the adp-ribosylating sirtuin (sirtm)2 from streptococcus pyogenes (apo form)
38 c2gb5B_
not modelled
41.0
24
PDB header: hydrolaseChain: B: PDB Molecule: nadh pyrophosphatase;PDBTitle: crystal structure of nadh pyrophosphatase (ec 3.6.1.22) (1790429) from2 escherichia coli k12 at 2.30 a resolution
39 c4c2mX_
not modelled
41.0
34
PDB header: transcriptionChain: X: PDB Molecule: dna-directed rna polymerase i subunit rpa12;PDBTitle: structure of rna polymerase i at 2.8 a resolution
40 c3cngC_
not modelled
40.5
37
PDB header: hydrolaseChain: C: PDB Molecule: nudix hydrolase;PDBTitle: crystal structure of nudix hydrolase from nitrosomonas europaea
41 c3j21i_
not modelled
39.4
45
PDB header: ribosomeChain: I: PDB Molecule: 50s ribosomal protein l13p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
42 d1ubdc3
not modelled
39.2
33
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
43 d6paxa1
not modelled
38.1
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain
44 d1u86a1
not modelled
37.5
40
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
45 c2yu3A_
not modelled
36.8
18
PDB header: transcriptionChain: A: PDB Molecule: dna-directed rna polymerase iii 39 kdaPDBTitle: solution structure of the domain swapped wingedhelix in dna-2 directed rna polymerase iii 39 kda polypeptide
46 d2j0151
not modelled
36.6
71
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L32p
47 c3j3v0_
not modelled
36.5
43
PDB header: ribosomeChain: 0: PDB Molecule: 50s ribosomal protein l32;PDBTitle: atomic model of the immature 50s subunit from bacillus subtilis (state2 i-a)
48 c4y97H_
not modelled
36.4
34
PDB header: transferaseChain: H: PDB Molecule: dna polymerase alpha catalytic subunit;PDBTitle: crystal structure of human pol alpha b-subunit in complex with c-2 terminal domain of catalytic subunit
49 c2nb9A_
not modelled
35.9
23
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of zitp zinc finger
50 c2odxA_
not modelled
35.9
22
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c oxidase polypeptide iv;PDBTitle: solution structure of zn(ii)cox4
51 d1jj2y_
not modelled
34.9
34
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L37ae
52 d1m2ka_
not modelled
34.2
27
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Sir2 family of transcriptional regulators
53 c1yshD_
not modelled
34.1
30
PDB header: structural protein/rnaChain: D: PDB Molecule: ribosomal protein l37a;PDBTitle: localization and dynamic behavior of ribosomal protein l30e
54 d1a1ia1
not modelled
33.9
29
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
55 c2zkrz_
not modelled
33.8
27
PDB header: ribosomal protein/rnaChain: Z: PDB Molecule: e site t-rna;PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
56 d2zjrz1
not modelled
33.6
71
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L32p
57 c4rulA_
not modelled
33.4
30
PDB header: isomerase/dnaChain: A: PDB Molecule: dna topoisomerase 1;PDBTitle: crystal structure of full-length e.coli topoisomerase i in complex2 with ssdna
58 d1vqoz1
not modelled
32.9
33
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L37ae
59 c3glsC_
not modelled
32.8
27
PDB header: hydrolaseChain: C: PDB Molecule: nad-dependent deacetylase sirtuin-3,PDBTitle: crystal structure of human sirt3
60 d1yuza2
not modelled
32.7
36
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin
61 d1zyba1
not modelled
32.2
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like
62 d1aaya1
not modelled
31.5
29
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
63 c2l8nA_
not modelled
31.3
17
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional repressor cytr;PDBTitle: nmr structure of the cytidine repressor dna binding domain in presence2 of operator half-site dna
64 c5xonU_
not modelled
30.9
17
PDB header: transcription/rnaChain: U: PDB Molecule: general transcription elongation factor tfiis;PDBTitle: rna polymerase ii elongation complex bound with spt4/5 and tfiis
65 d1lkoa2
not modelled
30.8
27
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin
66 d1nnqa2
not modelled
30.4
36
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin
67 d1ubdc4
not modelled
30.3
31
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
68 d1tf3a2
not modelled
29.9
14
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
69 c1i3qI_
not modelled
29.8
17
PDB header: transcriptionChain: I: PDB Molecule: dna-directed rna polymerase ii 14.2kdPDBTitle: rna polymerase ii crystal form i at 3.1 a resolution
70 d2ey4e1
not modelled
29.6
27
Fold: Rubredoxin-likeSuperfamily: Nop10-like SnoRNPFamily: Nucleolar RNA-binding protein Nop10-like
71 c3hhgF_
not modelled
29.5
11
PDB header: transcription regulatorChain: F: PDB Molecule: transcriptional regulator, lysr family;PDBTitle: structure of crga, a lysr-type transcriptional regulator from2 neisseria meningitidis.
72 c2aklA_
not modelled
29.2
39
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phna-like protein pa0128;PDBTitle: solution structure for phn-a like protein pa0128 from2 pseudomonas aeruginosa
73 d2f4ma1
not modelled
29.0
13
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core
74 c1s1i9_
not modelled
28.8
17
PDB header: ribosomeChain: 9: PDB Molecule: 60s ribosomal protein l43;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1i, contains 60s subunit. the 40s4 ribosomal subunit is in file 1s1h.
75 c2qa4Z_
not modelled
28.0
33
PDB header: ribosomeChain: Z: PDB Molecule: 50s ribosomal protein l37ae;PDBTitle: a more complete structure of the the l7/l12 stalk of the2 haloarcula marismortui 50s large ribosomal subunit
76 d1imla1
not modelled
27.6
44
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain
77 d2dlqa2
not modelled
27.3
57
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
78 d2cona1
not modelled
27.1
25
Fold: Rubredoxin-likeSuperfamily: NOB1 zinc finger-likeFamily: NOB1 zinc finger-like
79 d1yc5a1
not modelled
26.9
27
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Sir2 family of transcriptional regulators
80 c3eswA_
not modelled
26.8
18
PDB header: hydrolaseChain: A: PDB Molecule: peptide-n(4)-(n-acetyl-beta-glucosaminyl)asparaginePDBTitle: complex of yeast pngase with glcnac2-iac.
81 d1ffkw_
not modelled
26.3
27
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L37ae
82 d2gaua1
not modelled
25.8
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like
83 c1nuiA_
not modelled
25.8
13
PDB header: replicationChain: A: PDB Molecule: dna primase/helicase;PDBTitle: crystal structure of the primase fragment of bacteriophage t7 primase-2 helicase protein
84 c1areA_
not modelled
25.8
22
PDB header: transcription regulationChain: A: PDB Molecule: yeast transcription factor adr1;PDBTitle: structures of dna-binding mutant zinc finger domains: implications for2 dna binding
85 d1tdza3
not modelled
25.5
29
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins
86 c5fo5A_
not modelled
25.3
11
PDB header: transcriptionChain: A: PDB Molecule: hth-type transcriptional regulator metr;PDBTitle: structure of the dna-binding domain of escherichia coli methionine2 biosynthesis regulator metr
87 d1u85a1
not modelled
24.9
44
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
88 c3iz5i_
not modelled
24.9
25
PDB header: ribosomeChain: I: PDB Molecule: 60s ribosomal protein l10 (l10e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
89 c3lsgD_
not modelled
24.9
15
PDB header: transcription regulatorChain: D: PDB Molecule: two-component response regulator yesn;PDBTitle: the crystal structure of the c-terminal domain of the two-2 component response regulator yesn from fusobacterium3 nucleatum subsp. nucleatum atcc 25586
90 c3j21d_
not modelled
24.5
33
PDB header: ribosomeChain: D: PDB Molecule: 50s ribosomal protein l4p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
91 d1x6ea2
not modelled
24.5
33
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
92 d2esna1
not modelled
24.4
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: LysR-like transcriptional regulators
93 c5y9sD_
not modelled
24.4
14
PDB header: transcriptionChain: D: PDB Molecule: vv2_1132;PDBTitle: crystal structure of vv2_1132, a lysr family transcriptional regulator
94 c3j39g_
not modelled
24.1
17
PDB header: ribosomeChain: G: PDB Molecule: 60s ribosomal protein l7a;PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
95 d1q1aa_
not modelled
23.7
18
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Sir2 family of transcriptional regulators
96 d2apob1
not modelled
23.6
27
Fold: Rubredoxin-likeSuperfamily: Nop10-like SnoRNPFamily: Nucleolar RNA-binding protein Nop10-like
97 c3izci_
not modelled
23.3
33
PDB header: ribosomeChain: I: PDB Molecule: 60s ribosomal protein rpl10 (l10e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
98 c2elpA_
not modelled
23.3
33
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 13th c2h2 zinc finger of human2 zinc finger protein 406
99 d1sp2a_
not modelled
23.2
29
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
100 c2m6nA_
not modelled
23.1
17
PDB header: cell cycleChain: A: PDB Molecule: f-box only protein 5;PDBTitle: 3d solution structure of emi1 (early mitotic inhibitor 1)
101 d2coha1
not modelled
23.0
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like
102 d2yt9a1
not modelled
22.9
38
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
103 c3jyw9_
not modelled
22.8
18
PDB header: ribosomeChain: 9: PDB Molecule: 60s ribosomal protein l43;PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
104 c6f40P_
not modelled
22.7
20
PDB header: transcriptionChain: P: PDB Molecule: dna-directed rna polymerase iii subunit rpc6;PDBTitle: rna polymerase iii open complex
105 c4b6ap_
not modelled
22.6
18
PDB header: ribosomeChain: P: PDB Molecule: 60s ribosomal protein l17-a;PDBTitle: cryo-em structure of the 60s ribosomal subunit in complex2 with arx1 and rei1
106 c3ispA_
not modelled
22.5
14
PDB header: transcriptionChain: A: PDB Molecule: hth-type transcriptional regulatorPDBTitle: crystal structure of argp from mycobacterium tuberculosis
107 d1a1ia2
not modelled
22.4
33
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
108 d1f2ig1
not modelled
22.4
29
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
109 d3e5ua1
not modelled
22.4
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like
110 c1xf7A_
not modelled
22.3
25
PDB header: transcriptionChain: A: PDB Molecule: wilms' tumor protein;PDBTitle: high resolution nmr structure of the wilms' tumor2 suppressor protein (wt1) finger 3
111 d1xf7a_
not modelled
22.3
25
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
112 d1a1ga1
not modelled
22.2
29
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
113 c4ce45_
not modelled
22.1
27
PDB header: ribosomeChain: 5: PDB Molecule: mrpl32;PDBTitle: 39s large subunit of the porcine mitochondrial ribosome
114 c3k1nB_
not modelled
21.9
8
PDB header: transcriptionChain: B: PDB Molecule: hth-type transcriptional regulator benm;PDBTitle: crystal structure of full-length benm
115 d2oz6a1
not modelled
21.7
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like
116 c1va1A_
not modelled
21.7
21
PDB header: transcriptionChain: A: PDB Molecule: transcription factor sp1;PDBTitle: solution structure of transcription factor sp1 dna binding2 domain (zinc finger 1)
117 c3m8jA_
not modelled
21.5
8
PDB header: transcriptionChain: A: PDB Molecule: focb protein;PDBTitle: crystal structure of e.coli focb at 1.4 a resolution
118 d2ct1a1
not modelled
21.5
50
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
119 c5z4yB_
not modelled
21.5
8
PDB header: dna binding proteinChain: B: PDB Molecule: cys regulon transcriptional activator;PDBTitle: crystal structure of pacysb ntd domain with space group p4
120 c5ol0B_
not modelled
21.4
15
PDB header: hydrolaseChain: B: PDB Molecule: putative silent information regulator 2,putative silentPDBTitle: structure of leishmania infantum silent information regulator 22 related protein 1 (lisir2rp1) in complex with acetylated p53 peptide