1 c5c0wJ_
59.8
21
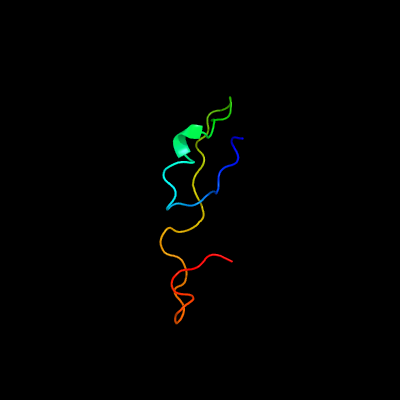
PDB header: hydrolase/rnaChain: J: PDB Molecule: exosome complex exonuclease dis3;PDBTitle: structure of a 12-subunit nuclear exosome complex bound to single-2 stranded rna substrates
2 c2wp8J_
57.0
21
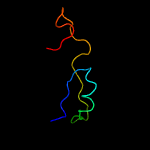
PDB header: hydrolaseChain: J: PDB Molecule: exosome complex exonuclease dis3;PDBTitle: yeast rrp44 nuclease
3 c6d6rK_
39.6
44
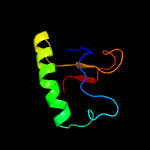
PDB header: hydrolaseChain: K: PDB Molecule: exosome complex exonuclease rrp44;PDBTitle: human nuclear exosome-mtr4 rna complex - composite map after focused2 reconstruction
4 c6md3F_
36.5
50
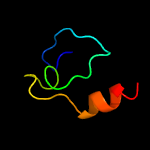
PDB header: hydrolase/rnaChain: F: PDB Molecule: rrp44p homologue;PDBTitle: structure of t. brucei rrp44 pin domain
5 d2a1ha1
32.5
18
Fold: D-aminoacid aminotransferase-like PLP-dependent enzymesSuperfamily: D-aminoacid aminotransferase-like PLP-dependent enzymesFamily: D-aminoacid aminotransferase-like PLP-dependent enzymes
6 c5oqj4_
27.9
25
PDB header: transcriptionChain: 4: PDB Molecule: rna polymerase ii transcription factor b subunit 4;PDBTitle: structure of yeast transcription pre-initiation complex with tfiih
7 c5n60Q_
27.0
50
PDB header: transferaseChain: Q: PDB Molecule: rna polymerase i-specific transcription initiation factorPDBTitle: cryo-em structure of rna polymerase i in complex with rrn3 and core2 factor (orientation i)
8 c2r9qD_
24.9
23
PDB header: hydrolaseChain: D: PDB Molecule: 2'-deoxycytidine 5'-triphosphate deaminase;PDBTitle: crystal structure of 2'-deoxycytidine 5'-triphosphate deaminase from2 agrobacterium tumefaciens
9 c2abjG_
22.0
18
PDB header: transferaseChain: G: PDB Molecule: branched-chain-amino-acid aminotransferase, cytosolic;PDBTitle: crystal structure of human branched chain amino acid transaminase in a2 complex with an inhibitor, c16h10n2o4f3scl, and pyridoxal 5'3 phosphate.
10 c6grvA_
19.2
22
PDB header: metal binding proteinChain: A: PDB Molecule: metallothionein;PDBTitle: cadmium(ii) form of full-length metallothionein from pseudomonas2 fluorescens q2-87 (pflq2 mt)
11 c5jcaS_
14.7
40
PDB header: oxidoreductaseChain: S: PDB Molecule: nadh-dependent ferredoxin:nadp oxidoreductase (nfni)PDBTitle: nadp(h) bound nadh-dependent ferredoxin:nadp oxidoreductase (nfni)2 from pyrococcus furiosus
12 c4ylfA_
14.2
40
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydroorotate dehydrogenase b (nad(+)), electron transferPDBTitle: insights into flavin-based electron bifurcation via the nadh-dependent2 reduced ferredoxin-nadp oxidoreductase structure
13 d1rfma_
13.5
30
Fold: L-sulfolactate dehydrogenase-likeSuperfamily: L-sulfolactate dehydrogenase-likeFamily: L-sulfolactate dehydrogenase-like
14 c5op0B_
13.0
13
PDB header: transferaseChain: B: PDB Molecule: dna polymerase ligd, polymerase domain;PDBTitle: structure of prim-polc from mycobacterium smegmatis
15 c2iruA_
12.2
10
PDB header: transferaseChain: A: PDB Molecule: putative dna ligase-like protein rv0938/mt0965;PDBTitle: crystal structure of the polymerase domain from mycobacterium2 tuberculosis ligase d
16 d1nxua_
11.7
14
Fold: L-sulfolactate dehydrogenase-likeSuperfamily: L-sulfolactate dehydrogenase-likeFamily: L-sulfolactate dehydrogenase-like
17 d1xrha_
11.6
19
Fold: L-sulfolactate dehydrogenase-likeSuperfamily: L-sulfolactate dehydrogenase-likeFamily: L-sulfolactate dehydrogenase-like
18 c3uoeB_
11.1
35
PDB header: oxidoreductaseChain: B: PDB Molecule: dehydrogenase;PDBTitle: the crystal structure of dehydrogenase from sinorhizobium meliloti
19 c1ep3B_
10.7
50
PDB header: oxidoreductaseChain: B: PDB Molecule: dihydroorotate dehydrogenase b (pyrk subunit);PDBTitle: crystal structure of lactococcus lactis dihydroorotate dehydrogenase2 b. data collected under cryogenic conditions.
20 c3lq9B_
10.6
44
PDB header: signaling proteinChain: B: PDB Molecule: dna-damage-inducible transcript 4 protein;PDBTitle: crystal structure of human redd1, a hypoxia-induced regulator of mtor
21 c6bymA_
not modelled
10.2
22
PDB header: lipid transportChain: A: PDB Molecule: sterol-binding protein;PDBTitle: crystal structure of the sterol-bound second start domain of yeast2 lam4
22 c5vj7B_
not modelled
8.7
40
PDB header: oxidoreductaseChain: B: PDB Molecule: ferredoxin-nadp(+) reductase subunit alpha;PDBTitle: ferredoxin nadp oxidoreductase (xfn)
23 c5dmuA_
not modelled
8.3
16
PDB header: transferaseChain: A: PDB Molecule: nhej polymerase;PDBTitle: structure of the nhej polymerase from methanocella paludicola
24 c2piaA_
not modelled
7.4
33
PDB header: reductaseChain: A: PDB Molecule: phthalate dioxygenase reductase;PDBTitle: phthalate dioxygenase reductase: a modular structure for electron2 transfer from pyridine nucleotides to [2fe-2s]
25 c1d4cB_
not modelled
7.2
24
PDB header: oxidoreductaseChain: B: PDB Molecule: flavocytochrome c fumarate reductase;PDBTitle: crystal structure of the uncomplexed form of the flavocytochrome c2 fumarate reductase of shewanella putrefaciens strain mr-1
26 c2kfqA_
not modelled
7.1
30
PDB header: de novo proteinChain: A: PDB Molecule: fp1;PDBTitle: nmr structure of fp1
27 c2n9oA_
not modelled
6.9
33
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase rnf126;PDBTitle: solution structure of rnf126 n-terminal zinc finger domain
28 d1ep3b2
not modelled
6.7
56
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: Dihydroorotate dehydrogenase B, PyrK subunit
29 c2faoB_
not modelled
6.7
19
PDB header: hydrolase/transferaseChain: B: PDB Molecule: probable atp-dependent dna ligase;PDBTitle: crystal structure of pseudomonas aeruginosa ligd polymerase2 domain
30 c2m2jA_
not modelled
6.4
32
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative periplasmic protein;PDBTitle: solution nmr structure of the n-terminal domain of stm1478 from2 salmonella typhimurium lt2: target str147a of the northeast3 structural genomics consortium (nesg), and apc101565 of the midwest4 center for structural genomics (mcsg).
31 d1awda_
not modelled
6.4
27
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related
32 c1j5mA_
not modelled
6.3
75
PDB header: metal binding proteinChain: A: PDB Molecule: metallothionein-1;PDBTitle: solution structure of the synthetic 113cd_3 beta_n domain2 of lobster metallothionein-1
33 d1frra_
not modelled
6.2
31
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related
34 c2g8yB_
not modelled
6.1
23
PDB header: oxidoreductaseChain: B: PDB Molecule: malate/l-lactate dehydrogenases;PDBTitle: the structure of a putative malate/lactate dehydrogenase from e. coli.
35 c3uzbA_
not modelled
5.8
17
PDB header: transferaseChain: A: PDB Molecule: branched-chain-amino-acid aminotransferase;PDBTitle: crystal structures of branched-chain aminotransferase from deinococcus2 radiodurans complexes with alpha-ketoisocaproate and l-glutamate3 suggest its radio-resistance for catalysis
36 d1fxia_
not modelled
5.8
33
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related
37 c1wtjB_
not modelled
5.6
36
PDB header: oxidoreductaseChain: B: PDB Molecule: ureidoglycolate dehydrogenase;PDBTitle: crystal structure of delta1-piperideine-2-carboxylate2 reductase from pseudomonas syringae pvar.tomato
38 d2axth1
not modelled
5.5
27
Fold: Single transmembrane helixSuperfamily: Photosystem II 10 kDa phosphoprotein PsbHFamily: PsbH-like
39 c2axtH_
not modelled
5.5
27
PDB header: electron transportChain: H: PDB Molecule: photosystem ii reaction center h protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus