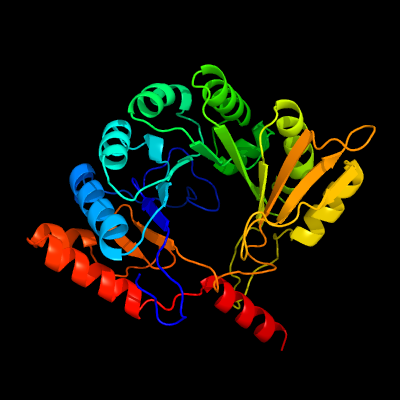
1 d1tv8a_
100.0
33
Fold: TIM beta/alpha-barrelSuperfamily: Radical SAM enzymesFamily: MoCo biosynthesis proteins
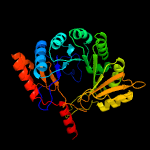
2 c6efnA_
100.0
19
PDB header: oxidoreductaseChain: A: PDB Molecule: sporulation killing factor maturation protein skfb;PDBTitle: structure of a ripp maturase, skfb
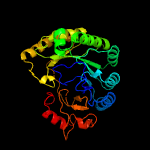
3 c5v1tA_
100.0
16
PDB header: metal binding proteinChain: A: PDB Molecule: radical sam;PDBTitle: crystal structure of streptococcus suis suib bound to precursor2 peptide suia
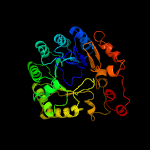
4 c4k39A_
100.0
19
PDB header: oxidoreductaseChain: A: PDB Molecule: anaerobic sulfatase-maturating enzyme;PDBTitle: native ansmecpe with bound adomet and cp18cys peptide
5 c5wggA_
100.0
13
PDB header: peptide binding proteinChain: A: PDB Molecule: radical sam domain protein;PDBTitle: structural insights into thioether bond formation in the biosynthesis2 of sactipeptides
6 c6c8vA_
100.0
22
PDB header: oxidoreductaseChain: A: PDB Molecule: coenzyme pqq synthesis protein e;PDBTitle: x-ray structure of pqqe from methylobacterium extorquens
7 c6b4cH_
100.0
15
PDB header: antiviral proteinChain: H: PDB Molecule: viperin;PDBTitle: structure of viperin from trichoderma virens
8 c4wcxC_
100.0
15
PDB header: lyaseChain: C: PDB Molecule: biotin and thiamin synthesis associated;PDBTitle: crystal structure of hydg: a maturase of the [fefe]-hydrogenase
9 c5vslB_
100.0
20
PDB header: antiviral proteinChain: B: PDB Molecule: radical s-adenosyl methionine domain-containing protein 2;PDBTitle: crystal structure of viperin with bound [4fe-4s] cluster and s-2 adenosylhomocysteine (sah)
10 c2yx0A_
99.9
14
PDB header: metal binding proteinChain: A: PDB Molecule: radical sam enzyme;PDBTitle: crystal structure of p. horikoshii tyw1
11 c4m7tA_
99.9
21
PDB header: metal binding proteinChain: A: PDB Molecule: btrn;PDBTitle: crystal structure of btrn in complex with adomet and 2-doia
12 c3c8fA_
99.9
13
PDB header: oxidoreductaseChain: A: PDB Molecule: pyruvate formate-lyase 1-activating enzyme;PDBTitle: 4fe-4s-pyruvate formate-lyase activating enzyme with partially2 disordered adomet
13 c5th5C_
99.9
14
PDB header: lyaseChain: C: PDB Molecule: 7-carboxy-7-deazaguanine synthase;PDBTitle: crystal structure of quee from bacillus subtilis with 6-carboxypterin-2 5'-deoxyadenosyl ester bound
14 c3t7vA_
99.8
16
PDB header: transferaseChain: A: PDB Molecule: methylornithine synthase pylb;PDBTitle: crystal structure of methylornithine synthase (pylb)
15 c3rfaB_
99.8
21
PDB header: oxidoreductaseChain: B: PDB Molecule: ribosomal rna large subunit methyltransferase n;PDBTitle: x-ray structure of rlmn from escherichia coli in complex with s-2 adenosylmethionine
16 d1r30a_
99.8
20
Fold: TIM beta/alpha-barrelSuperfamily: Radical SAM enzymesFamily: Biotin synthase
17 c1r30A_
99.8
20
PDB header: transferaseChain: A: PDB Molecule: biotin synthase;PDBTitle: the crystal structure of biotin synthase, an s-adenosylmethionine-2 dependent radical enzyme
18 c2a5hC_
99.8
19
PDB header: isomeraseChain: C: PDB Molecule: l-lysine 2,3-aminomutase;PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
19 c5exkG_
99.8
21
PDB header: transferaseChain: G: PDB Molecule: lipoyl synthase;PDBTitle: crystal structure of m. tuberculosis lipoyl synthase with 6-2 thiooctanoyl peptide intermediate
20 c3cixA_
99.8
17
PDB header: adomet binding proteinChain: A: PDB Molecule: fefe-hydrogenase maturase;PDBTitle: x-ray structure of the [fefe]-hydrogenase maturase hyde from2 thermotoga maritima in complex with thiocyanate
21 c6fz6B_
not modelled
99.8
18
PDB header: transferaseChain: B: PDB Molecule: probable dual-specificity rna methyltransferase rlmn;PDBTitle: crystal structure of a radical sam methyltransferase from2 sphaerobacter thermophilus
22 c3rfaA_
not modelled
99.8
20
PDB header: oxidoreductaseChain: A: PDB Molecule: ribosomal rna large subunit methyltransferase n;PDBTitle: x-ray structure of rlmn from escherichia coli in complex with s-2 adenosylmethionine
23 c4rtbA_
not modelled
99.8
14
PDB header: lyaseChain: A: PDB Molecule: hydg protein;PDBTitle: x-ray structure of the fefe-hydrogenase maturase hydg from2 carboxydothermus hydrogenoformans
24 c4jc0B_
not modelled
99.8
12
PDB header: transferaseChain: B: PDB Molecule: ribosomal protein s12 methylthiotransferase rimo;PDBTitle: crystal structure of thermotoga maritima holo rimo in complex with2 pentasulfide, northeast structural genomics consortium target vr77
25 c4u0pB_
not modelled
99.8
17
PDB header: transferaseChain: B: PDB Molecule: lipoyl synthase 2;PDBTitle: the crystal structure of lipoyl synthase in complex with s-adenosyl2 homocysteine
26 c2z2uA_
not modelled
99.7
17
PDB header: metal binding proteinChain: A: PDB Molecule: upf0026 protein mj0257;PDBTitle: crystal structure of archaeal tyw1
27 c3canA_
not modelled
99.7
14
PDB header: lyase activatorChain: A: PDB Molecule: pyruvate-formate lyase-activating enzyme;PDBTitle: crystal structure of a domain of pyruvate-formate lyase-activating2 enzyme from bacteroides vulgatus atcc 8482
28 c4njkA_
not modelled
99.7
23
PDB header: lyaseChain: A: PDB Molecule: 7-carboxy-7-deazaguanine synthase;PDBTitle: crystal structure of quee from burkholderia multivorans in complex2 with adomet, 7-carboxy-7-deazaguanine, and mg2+
29 c6nhlB_
not modelled
99.7
17
PDB header: lyaseChain: B: PDB Molecule: 7-carboxy-7-deazaguanine synthase;PDBTitle: crystal structure of quee from escherichia coli
30 d1olta_
not modelled
99.7
15
Fold: TIM beta/alpha-barrelSuperfamily: Radical SAM enzymesFamily: Oxygen-independent coproporphyrinogen III oxidase HemN
31 c6fd2B_
not modelled
99.6
19
PDB header: biosynthetic proteinChain: B: PDB Molecule: putative apramycin biosynthetic oxidoreductase 4;PDBTitle: radical sam 1,2-diol dehydratase aprd4 in complex with its substrate2 paromamine
32 c4r33A_
not modelled
99.6
18
PDB header: lyaseChain: A: PDB Molecule: nosl;PDBTitle: x-ray structure of the tryptophan lyase nosl with tryptophan and s-2 adenosyl-l-homocysteine bound
33 c5ul4A_
not modelled
99.5
15
PDB header: metal binding proteinChain: A: PDB Molecule: oxsb protein;PDBTitle: structure of cobalamin-dependent s-adenosylmethionine radical enzyme2 oxsb with aqua-cobalamin and s-adenosylmethionine bound
34 c5l7jA_
not modelled
99.3
15
PDB header: translationChain: A: PDB Molecule: elp3 family;PDBTitle: crystal structure of elp3 from dehalococcoides mccartyi
35 c2qgqF_
not modelled
99.3
12
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: protein tm_1862;PDBTitle: crystal structure of tm_1862 from thermotoga maritima. northeast2 structural genomics consortium target vr77
36 c6qk7C_
not modelled
99.1
16
PDB header: translationChain: C: PDB Molecule: elongator complex protein 3;PDBTitle: elongator catalytic subcomplex elp123 lobe
37 c4fheA_
not modelled
99.1
16
PDB header: lyaseChain: A: PDB Molecule: spore photoproduct lyase;PDBTitle: spore photoproduct lyase c140a mutant
38 c6iazA_
not modelled
98.9
13
PDB header: transferaseChain: A: PDB Molecule: histone acetyltransferase, elp3 family;PDBTitle: the archaeal methanocaldococcus infernus elp3 with n-terminus deletion2 (1-46)
39 c3ivuB_
not modelled
97.8
17
PDB header: transferaseChain: B: PDB Molecule: homocitrate synthase, mitochondrial;PDBTitle: homocitrate synthase lys4 bound to 2-og
40 c3rmjB_
not modelled
96.9
13
PDB header: transferaseChain: B: PDB Molecule: 2-isopropylmalate synthase;PDBTitle: crystal structure of truncated alpha-isopropylmalate synthase from2 neisseria meningitidis
41 c3bleA_
not modelled
96.8
13
PDB header: transferaseChain: A: PDB Molecule: citramalate synthase from leptospira interrogans;PDBTitle: crystal structure of the catalytic domain of licms in complexed with2 malonate
42 c3ewbX_
not modelled
96.7
14
PDB header: transferaseChain: X: PDB Molecule: 2-isopropylmalate synthase;PDBTitle: crystal structure of n-terminal domain of putative 2-isopropylmalate2 synthase from listeria monocytogenes
43 c6e1jB_
not modelled
96.6
12
PDB header: plant proteinChain: B: PDB Molecule: 2-isopropylmalate synthase, a genome specific 1;PDBTitle: crystal structure of methylthioalkylmalate synthase (bjumam1.1) from2 brassica juncea
44 c2ftpA_
not modelled
96.5
16
PDB header: lyaseChain: A: PDB Molecule: hydroxymethylglutaryl-coa lyase;PDBTitle: crystal structure of hydroxymethylglutaryl-coa lyase from pseudomonas2 aeruginosa
45 c3eegB_
not modelled
96.2
18
PDB header: transferaseChain: B: PDB Molecule: 2-isopropylmalate synthase;PDBTitle: crystal structure of a 2-isopropylmalate synthase from2 cytophaga hutchinsonii
46 c2cw6B_
not modelled
95.0
14
PDB header: lyaseChain: B: PDB Molecule: hydroxymethylglutaryl-coa lyase, mitochondrial;PDBTitle: crystal structure of human hmg-coa lyase: insights into2 catalysis and the molecular basis for3 hydroxymethylglutaric aciduria
47 c1ydoC_
not modelled
94.7
16
PDB header: lyaseChain: C: PDB Molecule: hmg-coa lyase;PDBTitle: crystal structure of the bacillis subtilis hmg-coa lyase, northeast2 structural genomics target sr181.
48 c1ydnA_
not modelled
92.4
17
PDB header: lyaseChain: A: PDB Molecule: hydroxymethylglutaryl-coa lyase;PDBTitle: crystal structure of the hmg-coa lyase from brucella melitensis,2 northeast structural genomics target lr35.
49 c1nvmG_
not modelled
92.1
17
PDB header: lyase/oxidoreductaseChain: G: PDB Molecule: 4-hydroxy-2-oxovalerate aldolase;PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
50 c4jn6C_
not modelled
91.9
19
PDB header: lyase/oxidoreductaseChain: C: PDB Molecule: 4-hydroxy-2-oxovalerate aldolase;PDBTitle: crystal structure of the aldolase-dehydrogenase complex from2 mycobacterium tuberculosis hrv37
51 d1nvma2
not modelled
91.6
15
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: HMGL-like
52 c5zmyF_
not modelled
90.1
14
PDB header: hydrolaseChain: F: PDB Molecule: cis-epoxysuccinate hydrolase;PDBTitle: crystal structure of a cis-epoxysuccinate hydrolase producing d(-)-2 tartaric acids
53 c4ov9A_
not modelled
90.1
11
PDB header: transferaseChain: A: PDB Molecule: isopropylmalate synthase;PDBTitle: structure of isopropylmalate synthase binding with alpha-2 isopropylmalate
54 c1rr2A_
not modelled
82.0
20
PDB header: transferaseChain: A: PDB Molecule: transcarboxylase 5s subunit;PDBTitle: propionibacterium shermanii transcarboxylase 5s subunit bound to 2-2 ketobutyric acid
55 c3hpxB_
not modelled
80.2
15
PDB header: transferaseChain: B: PDB Molecule: 2-isopropylmalate synthase;PDBTitle: crystal structure of mycobacterium tuberculosis leua active site2 domain 1-425 (truncation mutant delta:426-644)
56 c4lrtC_
not modelled
79.6
23
PDB header: lyase/oxidoreductaseChain: C: PDB Molecule: 4-hydroxy-2-oxovalerate aldolase;PDBTitle: crystal and solution structures of the bifunctional enzyme2 (aldolase/aldehyde dehydrogenase) from thermomonospora curvata,3 reveal a cofactor-binding domain motion during nad+ and coa4 accommodation whithin the shared cofactor-binding site
57 c3khdC_
not modelled
78.5
12
PDB header: transferaseChain: C: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of pff1300w.
58 c1sr9A_
not modelled
77.9
13
PDB header: transferaseChain: A: PDB Molecule: 2-isopropylmalate synthase;PDBTitle: crystal structure of leua from mycobacterium tuberculosis
59 c2zyfA_
not modelled
75.4
13
PDB header: transferaseChain: A: PDB Molecule: homocitrate synthase;PDBTitle: crystal structure of homocitrate synthase from thermus thermophilus2 complexed with magnesuim ion and alpha-ketoglutarate
60 c5xfmD_
not modelled
72.9
17
PDB header: hydrolaseChain: D: PDB Molecule: alpha-glucosidase;PDBTitle: crystal structure of beta-arabinopyranosidase
61 d1u7pa_
not modelled
72.3
11
Fold: HAD-likeSuperfamily: HAD-likeFamily: Magnesium-dependent phosphatase-1, Mdp1
62 c5ks8D_
not modelled
71.1
12
PDB header: ligaseChain: D: PDB Molecule: pyruvate carboxylase subunit beta;PDBTitle: crystal structure of two-subunit pyruvate carboxylase from2 methylobacillus flagellatus
63 c2iswB_
not modelled
70.9
21
PDB header: lyaseChain: B: PDB Molecule: putative fructose-1,6-bisphosphate aldolase;PDBTitle: structure of giardia fructose-1,6-biphosphate aldolase in2 complex with phosphoglycolohydroxamate
64 c3a9iA_
not modelled
64.1
13
PDB header: transferase/transferase inhibitorChain: A: PDB Molecule: homocitrate synthase;PDBTitle: crystal structure of homocitrate synthase from thermus thermophilus2 complexed with lys
65 c3chvA_
not modelled
63.3
13
PDB header: metal binding proteinChain: A: PDB Molecule: prokaryotic domain of unknown function (duf849) with a timPDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 member (spoa0042) from silicibacter pomeroyi dss-3 at 1.45 a3 resolution
66 c2zq0B_
not modelled
62.4
17
PDB header: hydrolaseChain: B: PDB Molecule: alpha-glucosidase (alpha-glucosidase susb);PDBTitle: crystal structure of susb complexed with acarbose
67 c1qhoA_
not modelled
61.5
21
PDB header: hydrolaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: five-domain alpha-amylase from bacillus stearothermophilus,2 maltose/acarbose complex
68 c5m99A_
not modelled
61.1
15
PDB header: hydrolaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: functional characterization and crystal structure of thermostable2 amylase from thermotoga petrophila, reveals high thermostability and3 an archaic form of dimerization
69 c2nx9B_
not modelled
59.6
15
PDB header: lyaseChain: B: PDB Molecule: oxaloacetate decarboxylase 2, subunit alpha;PDBTitle: crystal structure of the carboxyltransferase domain of the2 oxaloacetate decarboxylase na+ pump from vibrio cholerae
70 c3pm6B_
not modelled
59.2
13
PDB header: lyaseChain: B: PDB Molecule: putative fructose-bisphosphate aldolase;PDBTitle: crystal structure of a putative fructose-1,6-biphosphate aldolase from2 coccidioides immitis solved by combined sad mr
71 c6irtA_
not modelled
59.0
13
PDB header: membrane proteinChain: A: PDB Molecule: 4f2 cell-surface antigen heavy chain;PDBTitle: human lat1-4f2hc complex bound with bch
72 c5kzmA_
not modelled
58.7
14
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: crystal structure of tryptophan synthase alpha-beta chain complex from2 francisella tularensis
73 c2wcsA_
not modelled
57.2
17
PDB header: hydrolaseChain: A: PDB Molecule: alpha amylase, catalytic region;PDBTitle: crystal structure of debranching enzyme from nostoc2 punctiforme (npde)
74 c3c6cA_
not modelled
56.3
16
PDB header: hydrolaseChain: A: PDB Molecule: 3-keto-5-aminohexanoate cleavage enzyme;PDBTitle: crystal structure of a putative 3-keto-5-aminohexanoate cleavage2 enzyme (reut_c6226) from ralstonia eutropha jmp134 at 1.72 a3 resolution
75 c2aaaA_
not modelled
55.4
15
PDB header: glycosidaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: calcium binding in alpha-amylases: an x-ray diffraction study at 2.12 angstroms resolution of two enzymes from aspergillus
76 d1bf2a3
not modelled
55.3
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
77 c2p0oA_
not modelled
54.2
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein duf871;PDBTitle: crystal structure of a conserved protein from locus ef_2437 in2 enterococcus faecalis with an unknown function
78 d2nlya1
not modelled
53.9
11
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: Divergent polysaccharide deacetylase
79 c2lqoA_
not modelled
52.6
18
PDB header: oxidoreductaseChain: A: PDB Molecule: putative glutaredoxin rv3198.1/mt3292;PDBTitle: mrx1 reduced
80 c3dxiB_
not modelled
52.5
10
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative aldolase;PDBTitle: crystal structure of the n-terminal domain of a putative2 aldolase (bvu_2661) from bacteroides vulgatus
81 d1pama4
not modelled
52.1
10
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
82 c5h06C_
not modelled
51.2
18
PDB header: hydrolaseChain: C: PDB Molecule: amyp;PDBTitle: crystal structure of amyp in complex with maltose
83 d2obba1
not modelled
50.9
11
Fold: HAD-likeSuperfamily: HAD-likeFamily: BT0820-like
84 c6arhA_
not modelled
50.2
17
PDB header: lyaseChain: A: PDB Molecule: n-acetylneuraminate lyase;PDBTitle: crystal structure of human nal at a resolution of 1.6 angstrom
85 d1ea9c3
not modelled
48.8
24
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
86 c5hqcA_
not modelled
48.6
14
PDB header: hydrolaseChain: A: PDB Molecule: glycoside hydrolase family 97 enzyme;PDBTitle: a glycoside hydrolase family 97 enzyme r171k variant from2 pseudoalteromonas sp. strain k8
87 d1qhoa4
not modelled
47.8
25
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
88 d3bmva4
not modelled
47.8
13
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
89 c5d88A_
not modelled
47.5
15
PDB header: hydrolaseChain: A: PDB Molecule: predicted protease of the collagenase family;PDBTitle: the structure of the u32 peptidase mk0906
90 c5zxgB_
not modelled
46.0
20
PDB header: hydrolaseChain: B: PDB Molecule: cyclic maltosyl-maltose hydrolase;PDBTitle: cyclic alpha-maltosyl-(1-->6)-maltose hydrolase from arthrobacter2 globiformis, ligand-free form
91 c3a47A_
not modelled
45.5
19
PDB header: hydrolaseChain: A: PDB Molecule: oligo-1,6-glucosidase;PDBTitle: crystal structure of isomaltase from saccharomyces cerevisiae
92 c5zcbA_
not modelled
45.5
20
PDB header: hydrolaseChain: A: PDB Molecule: alpha-glucosidase;PDBTitle: crystal structure of alpha-glucosidase
93 d1cgta4
not modelled
44.1
11
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
94 c3vndD_
not modelled
43.9
11
PDB header: lyaseChain: D: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: crystal structure of tryptophan synthase alpha-subunit from the2 psychrophile shewanella frigidimarina k14-2
95 c4as2D_
not modelled
43.6
14
PDB header: hydrolaseChain: D: PDB Molecule: phosphorylcholine phosphatase;PDBTitle: pseudomonas aeruginosa phosphorylcholine phosphatase. monoclinic form
96 d1yhta1
not modelled
43.3
19
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-N-acetylhexosaminidase catalytic domain
97 d1gvia3
not modelled
43.0
22
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
98 c1cygA_
not modelled
42.9
17
PDB header: glycosyltransferaseChain: A: PDB Molecule: cyclodextrin glucanotransferase;PDBTitle: cyclodextrin glucanotransferase (e.c.2.4.1.19) (cgtase)
99 c3no5C_
not modelled
42.4
15
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a pfam duf849 domain containing protein2 (reut_a1631) from ralstonia eutropha jmp134 at 1.90 a resolution
100 d1ua7a2
not modelled
41.9
13
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
101 c1bagA_
not modelled
41.2
17
PDB header: alpha-amylaseChain: A: PDB Molecule: alpha-1,4-glucan-4-glucanohydrolase;PDBTitle: alpha-amylase from bacillus subtilis complexed with2 maltopentaose
102 d1n7ka_
not modelled
41.1
17
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
103 c3wy3A_
not modelled
40.6
17
PDB header: hydrolaseChain: A: PDB Molecule: alpha-glucosidase;PDBTitle: crystal structure of alpha-glucosidase mutant d202n in complex with2 glucose and glycerol
104 c1m53A_
not modelled
40.4
20
PDB header: isomeraseChain: A: PDB Molecule: isomaltulose synthase;PDBTitle: crystal structure of isomaltulose synthase (pali) from2 klebsiella sp. lx3
105 d1h3ga3
not modelled
40.4
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
106 c1lwhA_
not modelled
40.1
9
PDB header: transferaseChain: A: PDB Molecule: 4-alpha-glucanotransferase;PDBTitle: crystal structure of t. maritima 4-alpha-glucanotransferase
107 c2y7eA_
not modelled
39.4
18
PDB header: lyaseChain: A: PDB Molecule: 3-keto-5-aminohexanoate cleavage enzyme;PDBTitle: crystal structure of the 3-keto-5-aminohexanoate cleavage enzyme2 (kce) from candidatus cloacamonas acidaminovorans (tetragonal form)
108 d1uoka2
not modelled
39.0
26
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
109 c4jclA_
not modelled
38.1
13
PDB header: transferaseChain: A: PDB Molecule: cyclomaltodextrin glucanotransferase;PDBTitle: crystal structure of alpha-cgt from paenibacillus macerans at 1.72 angstrom resolution
110 c4j7rA_
not modelled
37.6
25
PDB header: hydrolaseChain: A: PDB Molecule: isoamylase;PDBTitle: crystal structure of chlamydomonas reinhardtii isoamylase 1 (isa1)
111 c4e2oA_
not modelled
37.6
22
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: alpha-amylase;PDBTitle: crystal structure of alpha-amylase from geobacillus thermoleovorans,2 gta, complexed with acarbose
112 d2gjpa2
not modelled
37.4
22
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
113 c5ey5A_
not modelled
37.3
19
PDB header: lyaseChain: A: PDB Molecule: lbcats-a;PDBTitle: lbcats
114 c3bmwA_
not modelled
37.2
11
PDB header: transferaseChain: A: PDB Molecule: cyclomaltodextrin glucanotransferase;PDBTitle: cyclodextrin glycosyl transferase from thermoanerobacterium2 thermosulfurigenes em1 mutant s77p complexed with a maltoheptaose3 inhibitor
115 d1m53a2
not modelled
37.0
22
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
116 d2aaaa2
not modelled
36.7
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
117 c6daoB_
not modelled
36.0
22
PDB header: lyaseChain: B: PDB Molecule: trans-o-hydroxybenzylidenepyruvate hydratase-aldolase;PDBTitle: nahe wt selenomethionine
118 d1e43a2
not modelled
36.0
11
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
119 c4n4qD_
not modelled
35.9
14
PDB header: lyaseChain: D: PDB Molecule: acylneuraminate lyase;PDBTitle: crystal structure of n-acetylneuraminate lyase from mycoplasma2 synoviae, crystal form ii
120 c5x7uA_
not modelled
35.5
20
PDB header: hydrolaseChain: A: PDB Molecule: trehalose synthase;PDBTitle: trehalose synthase from thermobaculum terrenum