| 1 |
|
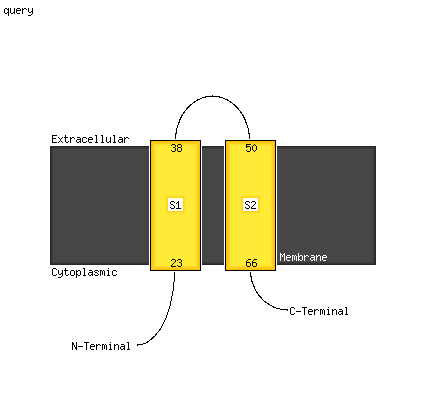
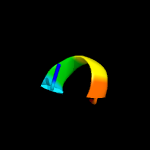
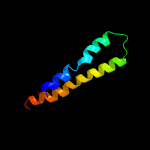
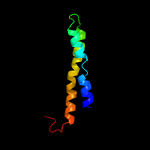
PDB 6adq chain C
Region: 7 - 45
Aligned: 39
Modelled: 39
Confidence: 21.6%
Identity: 23%
PDB header:electron transport
Chain: C: PDB Molecule:cytochrome bc1 complex cytochrome c subunit;
PDBTitle: respiratory complex ciii2civ2sod2 from mycobacterium smegmatis
Phyre2
| 2 |
|
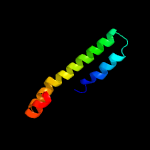
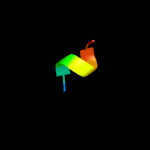
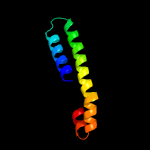
PDB 2kto chain A
Region: 50 - 60
Aligned: 11
Modelled: 11
Confidence: 18.7%
Identity: 27%
PDB header:antibiotic
Chain: A: PDB Molecule:lchb;
PDBTitle: spatial structure of lch-beta peptide from two-component lantibiotic2 lichenicidin vk21
Phyre2
| 3 |
|
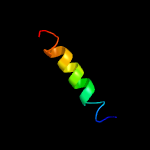
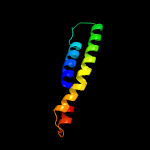
PDB 2klu chain A
Region: 60 - 75
Aligned: 16
Modelled: 16
Confidence: 11.7%
Identity: 31%
PDB header:immune system, membrane protein
Chain: A: PDB Molecule:t-cell surface glycoprotein cd4;
PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
Phyre2
| 4 |
|
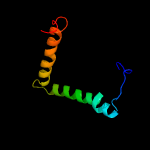
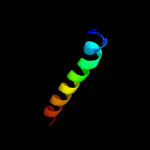
PDB 5f5v chain C
Region: 66 - 71
Aligned: 6
Modelled: 6
Confidence: 9.3%
Identity: 50%
PDB header:splicing
Chain: C: PDB Molecule:zinc finger domain-containing protein;
PDBTitle: crystal structure of the snu23-prp38-mfap1(217-296) complex of2 chaetomium thermophilum
Phyre2
| 5 |
|
PDB 5f5v chain F
Region: 66 - 71
Aligned: 6
Modelled: 6
Confidence: 9.3%
Identity: 50%
PDB header:splicing
Chain: F: PDB Molecule:zinc finger domain-containing protein;
PDBTitle: crystal structure of the snu23-prp38-mfap1(217-296) complex of2 chaetomium thermophilum
Phyre2
| 6 |
|
PDB 1uaz chain A
Region: 25 - 86
Aligned: 62
Modelled: 62
Confidence: 8.0%
Identity: 15%
Fold: Family A G protein-coupled receptor-like
Superfamily: Family A G protein-coupled receptor-like
Family: Bacteriorhodopsin-like
Phyre2
| 7 |
|
PDB 2lr1 chain B
Region: 66 - 71
Aligned: 6
Modelled: 6
Confidence: 7.9%
Identity: 50%
PDB header:apoptosis/signaling protein
Chain: B: PDB Molecule:immediate early glycoprotein;
PDBTitle: structural mechanism for bax inhibition by cytomegalovirus protein2 vmia
Phyre2
| 8 |
|
PDB 2low chain A
Region: 56 - 78
Aligned: 23
Modelled: 23
Confidence: 7.9%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:apelin receptor;
PDBTitle: solution structure of ar55 in 50% hfip
Phyre2
| 9 |
|
PDB 6f2w chain A
Region: 9 - 77
Aligned: 64
Modelled: 69
Confidence: 7.6%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:putative amino acid/polyamine transport protein;
PDBTitle: bacterial asc transporter crystal structure in open to in conformation
Phyre2
| 10 |
|
PDB 4iu8 chain A
Region: 20 - 84
Aligned: 65
Modelled: 65
Confidence: 7.5%
Identity: 6%
PDB header:transport protein
Chain: A: PDB Molecule:nitrite extrusion protein 2;
PDBTitle: crystal structure of a membrane transporter (selenomethionine2 derivative)
Phyre2
| 11 |
|
PDB 3ech chain C
Region: 70 - 77
Aligned: 8
Modelled: 8
Confidence: 7.3%
Identity: 63%
PDB header:transcription, transcription regulation
Chain: C: PDB Molecule:25-mer fragment of protein armr;
PDBTitle: the marr-family repressor mexr in complex with its antirepressor armr
Phyre2
| 12 |
|
PDB 5d6s chain B
Region: 9 - 35
Aligned: 27
Modelled: 27
Confidence: 7.3%
Identity: 11%
PDB header:oxidoreductase
Chain: B: PDB Molecule:epoxyqueuosine reductase;
PDBTitle: structure of epoxyqueuosine reductase from streptococcus thermophilus.
Phyre2
| 13 |
|
PDB 4iky chain A
Region: 20 - 87
Aligned: 68
Modelled: 68
Confidence: 7.2%
Identity: 18%
PDB header:transport protein
Chain: A: PDB Molecule:di-tripeptide abc transporter (permease);
PDBTitle: crystal structure of peptide transporter pot (e310q mutant) in complex2 with sulfate
Phyre2
| 14 |
|
PDB 2ifo chain A
Region: 54 - 72
Aligned: 19
Modelled: 19
Confidence: 7.1%
Identity: 21%
PDB header:virus
Chain: A: PDB Molecule:inovirus;
PDBTitle: model-building studies of inovirus: genetic variations on a2 geometric theme
Phyre2
| 15 |
|
PDB 2ww9 chain B
Region: 52 - 65
Aligned: 14
Modelled: 14
Confidence: 7.1%
Identity: 36%
PDB header:ribosome
Chain: B: PDB Molecule:protein transport protein sss1;
PDBTitle: cryo-em structure of the active yeast ssh1 complex bound to the yeast2 80s ribosome
Phyre2
| 16 |
|
PDB 1q90 chain D
Region: 12 - 39
Aligned: 28
Modelled: 28
Confidence: 6.7%
Identity: 29%
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 17 |
|
PDB 4hyj chain B
Region: 46 - 75
Aligned: 30
Modelled: 30
Confidence: 6.6%
Identity: 27%
PDB header:proton transport
Chain: B: PDB Molecule:rhodopsin;
PDBTitle: crystal structure of exiguobacterium sibiricum rhodopsin
Phyre2
| 18 |
|
PDB 2h3o chain A
Region: 60 - 71
Aligned: 12
Modelled: 12
Confidence: 5.7%
Identity: 33%
PDB header:membrane protein
Chain: A: PDB Molecule:merf;
PDBTitle: structure of merft, a membrane protein with two trans-2 membrane helices
Phyre2
| 19 |
|
PDB 4iu9 chain A
Region: 20 - 84
Aligned: 65
Modelled: 65
Confidence: 5.7%
Identity: 6%
PDB header:transport protein
Chain: A: PDB Molecule:nitrite extrusion protein 2;
PDBTitle: crystal structure of a membrane transporter
Phyre2
| 20 |
|
PDB 6gyh chain A
Region: 25 - 86
Aligned: 62
Modelled: 62
Confidence: 5.7%
Identity: 11%
PDB header:proton transport
Chain: A: PDB Molecule:family a g protein-coupled receptor-like protein;
PDBTitle: crystal structure of the light-driven proton pump coccomyxa2 subellipsoidea rhodopsin csr
Phyre2
| 21 |
|
| 22 |
|