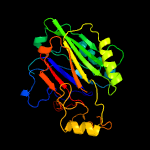
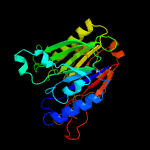
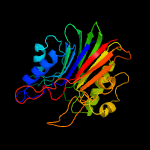
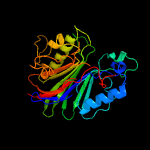
1 c5uvgA_
100.0
17
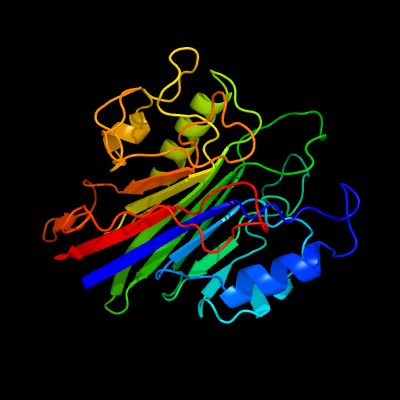
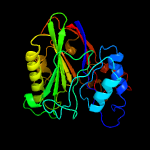
PDB header: hydrolaseChain: A: PDB Molecule: sphingomyelin phosphodiesterase 3,sphingomyelinPDBTitle: crystal structure of the human neutral sphingomyelinase 2 (nsmase2)2 catalytic domain with insertion deleted and calcium bound
2 c4ruwA_
99.9
19
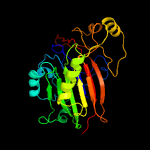
PDB header: hydrolaseChain: A: PDB Molecule: endonuclease/exonuclease/phosphatase;PDBTitle: the crystal structure of endonuclease/exonuclease/phosphatase from2 beutenbergia cavernae dsm 12333
3 c4f1iA_
99.9
19
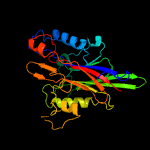
PDB header: hydrolaseChain: A: PDB Molecule: 5'-tyrosyl-dna phosphodiesterase;PDBTitle: crystal structure of semet tdp2 from caenorhabditis elegans
4 d1zwxa1
99.9
16
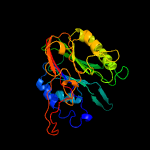
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: Sphingomyelin phosphodiesterase-like
5 c3wcxA_
99.9
20
PDB header: hydrolaseChain: A: PDB Molecule: sphingomyelinase c;PDBTitle: crystal structure of sphingomyelinase c from streptomyces2 griseocarneus
6 c4f1hA_
99.9
19
PDB header: hydrolase/dnaChain: A: PDB Molecule: tyrosyl-dna phosphodiesterase 2;PDBTitle: crystal structure of tdp2 from danio rerio complexed with a single2 strand dna
7 d2imqx1
99.9
17
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: Inositol polyphosphate 5-phosphatase (IPP5)
8 c3tebA_
99.9
18
PDB header: hydrolaseChain: A: PDB Molecule: endonuclease/exonuclease/phosphatase;PDBTitle: endonuclease/exonuclease/phosphatase family protein from leptotrichia2 buccalis c-1013-b
9 d2ddra1
99.9
16
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: Sphingomyelin phosphodiesterase-like
10 c3i46B_
99.9
17
PDB header: toxinChain: B: PDB Molecule: beta-hemolysin;PDBTitle: crystal structure of beta toxin from staphylococcus aureus f277a,2 p278a mutant with bound calcium ions
11 c3mprB_
99.9
14
PDB header: hydrolaseChain: B: PDB Molecule: putative endonuclease/exonuclease/phosphatase familyPDBTitle: crystal structure of endonuclease/exonuclease/phosphatase family2 protein from bacteroides thetaiotaomicron, northeast structural3 genomics consortium target btr318a
12 c3mtcA_
99.9
19
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: type ii inositol-1,4,5-trisphosphate 5-phosphatase;PDBTitle: crystal structure of inpp5b in complex with phosphatidylinositol 4-2 phosphate
13 c4fvaD_
99.9
18
PDB header: hydrolaseChain: D: PDB Molecule: 5'-tyrosyl-dna phosphodiesterase;PDBTitle: crystal structure of truncated caenorhabditis elegans tdp2
14 c4gz1B_
99.9
18
PDB header: hydrolase/dnaChain: B: PDB Molecule: tyrosyl-dna phosphodiesterase 2;PDBTitle: mus musculus tdp2 reaction product (5'-phosphorylated dna)-mg2+2 complex at 1.5 angstroms resolution
15 c6bt2B_
99.9
17
PDB header: hydrolase, rna binding proteinChain: B: PDB Molecule: nocturnin;PDBTitle: structure of the human nocturnin catalytic domain with bound sulfate2 anion
16 c3g6sA_
99.9
16
PDB header: hydrolaseChain: A: PDB Molecule: putative endonuclease/exonuclease/phosphatase familyPDBTitle: crystal structure of the endonuclease/exonuclease/phosphatase2 (bvu_0621) from bacteroides vulgatus. northeast structural genomics3 consortium target bvr56d
17 c3nr8A_
99.9
17
PDB header: hydrolaseChain: A: PDB Molecule: phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase 2;PDBTitle: crystal structure of human ship2
18 c3ngoA_
99.9
13
PDB header: hydrolase/dnaChain: A: PDB Molecule: ccr4-not transcription complex subunit 6-like;PDBTitle: crystal structure of the human cnot6l nuclease domain in complex with2 poly(a) dna
19 c3l1wE_
99.9
15
PDB header: structural genomics, unknown functionChain: E: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of a functionally unknown conserved2 protein from enterococcus faecalis v583
20 d1sr4b_
99.9
15
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: DNase I-like
21 c2j63B_
not modelled
99.9
17
PDB header: lyaseChain: B: PDB Molecule: ap-endonuclease;PDBTitle: crystal structure of ap endonuclease lmap from leishmania major
22 c4zkfA_
not modelled
99.9
19
PDB header: hydrolaseChain: A: PDB Molecule: 2',5'-phosphodiesterase 12;PDBTitle: crystal structure of human phosphodiesterase 12
23 d2f1na1
not modelled
99.9
17
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: DNase I-like
24 d2a40b1
not modelled
99.9
16
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: DNase I-like
25 c5j8nA_
not modelled
99.9
16
PDB header: hydrolaseChain: A: PDB Molecule: exodeoxyribonuclease iii;PDBTitle: exonuclease iii homologue mm3148 from methanosarcina mazei
26 c4k6lF_
not modelled
99.8
18
PDB header: toxinChain: F: PDB Molecule: cytolethal distending toxin subunit b homolog;PDBTitle: structure of typhoid toxin
27 c2jc5A_
not modelled
99.8
19
PDB header: hydrolaseChain: A: PDB Molecule: exodeoxyribonuclease;PDBTitle: apurinic apyrimidinic (ap) endonuclease (nape) from neisseria2 meningitidis
28 d1akoa_
not modelled
99.8
18
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: DNase I-like
29 c5ewtA_
not modelled
99.8
17
PDB header: hydrolaseChain: A: PDB Molecule: exodeoxyribonuclease iii xth;PDBTitle: crystal structure of exoiii endonuclease from sulfolobus islandicus
30 c1e9nB_
not modelled
99.8
15
PDB header: dna repairChain: B: PDB Molecule: dna-(apurinic or apyrimidinic site) lyase;PDBTitle: a second divalent metal ion in the active site of a new crystal form2 of human apurinic/apyrimidinic endonuclease, ape1, and its3 implications for the catalytic mechanism
31 c2jc4A_
not modelled
99.8
18
PDB header: hydrolaseChain: A: PDB Molecule: exodeoxyribonuclease iii;PDBTitle: 3'-5' exonuclease (nexo) from neisseria meningitidis
32 c4f1rA_
not modelled
99.8
15
PDB header: transcription regulatorChain: A: PDB Molecule: catabolite repression control protein;PDBTitle: structure analysis of the global metabolic regulator crc from2 pseudomonas aeruginos
33 c2xswB_
not modelled
99.8
18
PDB header: hydrolaseChain: B: PDB Molecule: 72 kda inositol polyphosphate 5-phosphatase;PDBTitle: crystal structure of human inpp5e
34 c2voaB_
not modelled
99.8
15
PDB header: lyaseChain: B: PDB Molecule: exodeoxyribonuclease iii;PDBTitle: structure of an ap endonuclease from archaeoglobus fulgidus
35 d1vyba_
not modelled
99.7
18
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: DNase I-like
36 c3g0rA_
not modelled
99.7
16
PDB header: hydrolase/dnaChain: A: PDB Molecule: exodeoxyribonuclease;PDBTitle: complex of mth0212 and an 8bp dsdna with distorted ends
37 c5cfeA_
not modelled
99.7
15
PDB header: hydrolaseChain: A: PDB Molecule: exodeoxyribonuclease;PDBTitle: bacillus subtilis ap endonuclease exoa
38 d1hd7a_
not modelled
99.7
16
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: DNase I-like
39 c4cmnA_
not modelled
99.6
17
PDB header: hydrolaseChain: A: PDB Molecule: inositol polyphosphate 5-phosphatase ocrl-1;PDBTitle: crystal structure of ocrl in complex with a phosphate ion
40 d1wdua_
not modelled
99.5
12
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: DNase I-like
41 c2ei9A_
not modelled
99.5
17
PDB header: gene regulationChain: A: PDB Molecule: non-ltr retrotransposon r1bmks orf2 protein;PDBTitle: crystal structure of r1bm endonuclease domain
42 c6ibdA_
not modelled
99.5
21
PDB header: hydrolaseChain: A: PDB Molecule: phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1;PDBTitle: the phosphatase and c2 domains of human ship1
43 d1i9za_
not modelled
99.5
18
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: Inositol polyphosphate 5-phosphatase (IPP5)
44 c5okmA_
not modelled
99.5
18
PDB header: hydrolaseChain: A: PDB Molecule: phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2;PDBTitle: crystal structure of human ship2 phosphatase-c2
45 c6a42A_
not modelled
98.5
19
PDB header: dna binding proteinChain: A: PDB Molecule: rna-directed dna polymerase homolog (r1),polyubiquitin-c;PDBTitle: r1en(5-223)-ubiquitin fusion
46 c2yhgA_
not modelled
82.5
19
PDB header: hydrolaseChain: A: PDB Molecule: cellulose-binding protein;PDBTitle: ab initio phasing of a nucleoside hydrolase-related hypothetical2 protein from saccharophagus degradans that is associated with3 carbohydrate metabolism
47 c6ihbR_
not modelled
73.9
21
PDB header: virusChain: R: PDB Molecule: dyslexia-associated protein kiaa0319-like protein;PDBTitle: adeno-associated virus 2 in complex with aavr
48 c2q8uA_
not modelled
70.4
15
PDB header: hydrolaseChain: A: PDB Molecule: exonuclease, putative;PDBTitle: crystal structure of mre11 from thermotoga maritima msb8 (tm1635) at2 2.20 a resolution
49 c3av0A_
not modelled
64.7
14
PDB header: recombinationChain: A: PDB Molecule: dna double-strand break repair protein mre11;PDBTitle: crystal structure of mre11-rad50 bound to atp s
50 c5iryB_
not modelled
62.6
14
PDB header: cell adhesionChain: B: PDB Molecule: desmocollin-1;PDBTitle: crystal structure of human desmocollin-1 ectodomain
51 c3ib7A_
not modelled
62.2
18
PDB header: hydrolaseChain: A: PDB Molecule: icc protein;PDBTitle: crystal structure of full length rv0805
52 c3mbfA_
not modelled
57.2
42
PDB header: lyaseChain: A: PDB Molecule: fructose-bisphosphate aldolase;PDBTitle: crystal structure of fructose bisphosphate aldolase from2 encephalitozoon cuniculi, bound to fructose 1,6-bisphosphate
53 d2hy1a1
not modelled
55.2
18
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: GpdQ-like
54 c2hy1A_
not modelled
55.2
18
PDB header: hydrolaseChain: A: PDB Molecule: rv0805;PDBTitle: crystal structure of rv0805
55 c5wlyA_
not modelled
54.0
23
PDB header: hydrolaseChain: A: PDB Molecule: udp-2,3-diacylglucosamine hydrolase;PDBTitle: e. coli lpxh- 8 mutations
56 c5erdB_
not modelled
52.4
16
PDB header: cell adhesionChain: B: PDB Molecule: desmoglein-2;PDBTitle: crystal structure of human desmoglein-2 ectodomain
57 d1ozha1
not modelled
52.2
21
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
58 d1jifa_
not modelled
47.3
26
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Antibiotic resistance proteins
59 c5d8mA_
not modelled
45.0
13
PDB header: hydrolaseChain: A: PDB Molecule: metagenomic carboxyl esterase mgs0156;PDBTitle: crystal structure of the metagenomic carboxyl esterase mgs0156
60 c5k8kA_
not modelled
44.9
16
PDB header: hydrolaseChain: A: PDB Molecule: udp-2,3-diacylglucosamine hydrolase;PDBTitle: structure of the haemophilus influenzae lpxh-lipid x complex
61 c6nvoA_
not modelled
44.7
22
PDB header: dna binding proteinChain: A: PDB Molecule: nuclease mpe;PDBTitle: crystal structure of pseudomonas putida nuclease mpe
62 c6dnsA_
not modelled
44.3
27
PDB header: hydrolaseChain: A: PDB Molecule: alpha-1,4-endofucoidanase;PDBTitle: endo-fucoidan hydrolase mffcna9 from glycoside hydrolase family 107
63 c4n04B_
not modelled
43.0
33
PDB header: oxidoreductaseChain: B: PDB Molecule: glyoxalase/bleomycin resistance protein/dioxygenase;PDBTitle: the crystal structure of glyoxalase / bleomycin resistance protein2 from catenulispora acidiphila dsm 44928
64 c3auzA_
not modelled
40.4
14
PDB header: recombinationChain: A: PDB Molecule: dna double-strand break repair protein mre11;PDBTitle: crystal structure of mre11 with manganese
65 c3qfnA_
not modelled
40.2
17
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of streptococcal asymmetric ap4a hydrolase and2 phosphodiesterase spr1479/saph in complex with inorganic phosphate
66 c6a4xA_
not modelled
39.1
33
PDB header: biosynthetic proteinChain: A: PDB Molecule: bleomycin resistance protein;PDBTitle: oxidase chap-h2
67 c4tvvA_
not modelled
38.8
17
PDB header: hydrolaseChain: A: PDB Molecule: tyrosine phosphatase ii superfamily protein;PDBTitle: crystal structure of lppa from legionella pneumophila
68 c4fcxB_
not modelled
38.8
12
PDB header: hydrolaseChain: B: PDB Molecule: dna repair protein rad32;PDBTitle: s.pombe mre11 apoenzym
69 c3cf4G_
not modelled
38.5
17
PDB header: oxidoreductaseChain: G: PDB Molecule: acetyl-coa decarboxylase/synthase epsilon subunit;PDBTitle: structure of the codh component of the m. barkeri acds complex
70 c3kx6C_
not modelled
37.6
47
PDB header: lyaseChain: C: PDB Molecule: fructose-bisphosphate aldolase;PDBTitle: crystal structure of fructose-1,6-bisphosphate aldolase from babesia2 bovis at 2.1a resolution
71 d1twua_
not modelled
37.0
47
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Hypothetical protein YycE
72 d1zpda1
not modelled
36.6
16
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
73 c1l3wA_
not modelled
35.6
15
PDB header: cell adhesion, metal binding proteinChain: A: PDB Molecule: ep-cadherin;PDBTitle: c-cadherin ectodomain
74 d2ji7a1
not modelled
35.1
26
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
75 c4m02A_
not modelled
35.0
20
PDB header: calcium binding proteinChain: A: PDB Molecule: serine-rich adhesin for platelets;PDBTitle: middle fragment(residues 494-663) of the binding region of srap
76 d2yvta1
not modelled
32.7
9
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: TT1561-like
77 d1ii7a_
not modelled
32.7
10
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: DNA double-strand break repair nuclease
78 c6a4zA_
not modelled
30.9
31
PDB header: biosynthetic proteinChain: A: PDB Molecule: chap protein;PDBTitle: oxidase chap
79 c5kinC_
not modelled
30.2
5
PDB header: lyaseChain: C: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: crystal structure of tryptophan synthase alpha beta complex from2 streptococcus pneumoniae
80 c3ff7B_
not modelled
29.6
13
PDB header: cell adhesion/immune systemChain: B: PDB Molecule: epithelial cadherin;PDBTitle: structure of nk cell receptor klrg1 bound to e-cadherin
81 d1pvda1
not modelled
28.9
8
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
82 c4ltyD_
not modelled
28.4
18
PDB header: hydrolaseChain: D: PDB Molecule: exonuclease subunit sbcd;PDBTitle: crystal structure of e.coli sbcd at 1.8 a resolution
83 c3t1iC_
not modelled
27.8
15
PDB header: hydrolaseChain: C: PDB Molecule: double-strand break repair protein mre11a;PDBTitle: crystal structure of human mre11: understanding tumorigenic mutations
84 c4k6lC_
not modelled
27.5
14
PDB header: toxinChain: C: PDB Molecule: putative pertussis-like toxin subunit;PDBTitle: structure of typhoid toxin
85 c2yopB_
not modelled
25.8
12
PDB header: apoptosisChain: B: PDB Molecule: protein fam3b;PDBTitle: long wavelength s-sad structure of fam3b pander
86 d2ihta1
not modelled
25.4
23
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
87 c5svan_
not modelled
25.3
32
PDB header: transcription, transferase/dnaChain: N: PDB Molecule: mediator of rna polymerase ii transcription subunit 8;PDBTitle: mediator-rna polymerase ii pre-initiation complex
88 d1ovma1
not modelled
24.8
11
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
89 c4ykeA_
not modelled
23.9
10
PDB header: hydrolaseChain: A: PDB Molecule: mre11;PDBTitle: crystal structure of eukaryotic mre11 catalytic domain from chaetomium2 thermophilum
90 d1a5ca_
not modelled
23.4
75
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
91 c4fbkB_
not modelled
23.0
12
PDB header: hydrolase, protein bindingChain: B: PDB Molecule: dna repair and telomere maintenance protein nbs1,dna repairPDBTitle: crystal structure of a covalently fused nbs1-mre11 complex with one2 manganese ion per active site
92 c2zw7A_
not modelled
22.5
28
PDB header: transferaseChain: A: PDB Molecule: bleomycin acetyltransferase;PDBTitle: crystal structure of bleomycin n-acetyltransferase complexed2 with bleomycin a2 and coenzyme a
93 d1zaia1
not modelled
22.5
75
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
94 d1qo5b_
not modelled
22.4
88
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
95 d1fdja_
not modelled
22.4
88
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
96 d2qapa1
not modelled
21.8
75
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
97 c2qapC_
not modelled
21.7
75
PDB header: lyaseChain: C: PDB Molecule: fructose-1,6-bisphosphate aldolase;PDBTitle: fructose-1,6-bisphosphate aldolase from leishmania mexicana
98 d1fbaa_
not modelled
21.4
50
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
99 c3qg5D_
not modelled
21.2
16
PDB header: hydrolaseChain: D: PDB Molecule: mre11;PDBTitle: the mre11:rad50 complex forms an atp dependent molecular clamp in dna2 double-strand break repair
100 c5b4bB_
not modelled
21.1
16
PDB header: hydrolaseChain: B: PDB Molecule: udp-2,3-diacylglucosamine hydrolase;PDBTitle: crystal structure of lpxh with lipid x in spacegroup c2
101 d1t9ba1
not modelled
20.4
21
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
102 d3d03a1
not modelled
20.2
19
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: GpdQ-like
103 c2iqtA_
not modelled
20.1
75
PDB header: lyaseChain: A: PDB Molecule: fructose-bisphosphate aldolase class 1;PDBTitle: crystal structure of fructose-bisphosphate aldolase, class i from2 porphyromonas gingivalis