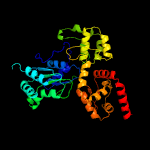
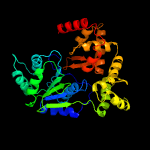
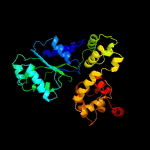
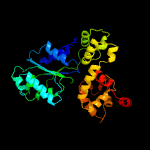
1 c1z6tC_
100.0
17
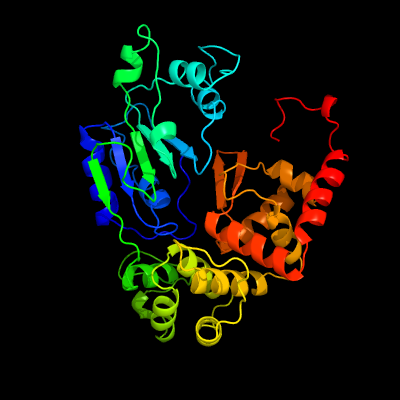
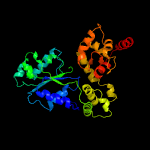
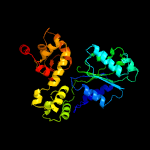
PDB header: apoptosisChain: C: PDB Molecule: apoptotic protease activating factor 1;PDBTitle: structure of the apoptotic protease-activating factor 12 bound to adp
2 c3iytG_
100.0
16
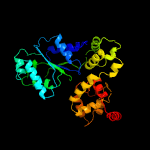
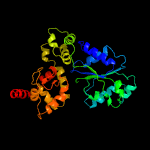
PDB header: apoptosisChain: G: PDB Molecule: apoptotic protease-activating factor 1;PDBTitle: structure of an apoptosome-procaspase-9 card complex
3 c2a5yB_
100.0
14
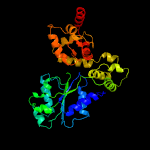
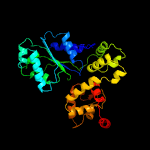
PDB header: apoptosisChain: B: PDB Molecule: ced-4;PDBTitle: structure of a ced-4/ced-9 complex
4 c5juyB_
100.0
17
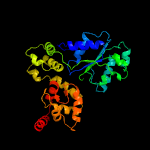
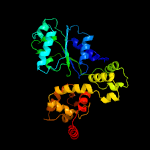
PDB header: apoptosisChain: B: PDB Molecule: apoptotic protease-activating factor 1;PDBTitle: active human apoptosome with procaspase-9
5 c6mfvC_
100.0
19
PDB header: signaling proteinChain: C: PDB Molecule: tetratricopeptide repeat sensor ph0952;PDBTitle: crystal structure of the signal transduction atpase with numerous2 domains (stand) protein with a tetratricopeptide repeat sensor ph09523 from pyrococcus horikoshii
6 c3iz8F_
100.0
17
PDB header: apoptosisChain: F: PDB Molecule: apaf-1 related killer dark;PDBTitle: structure of the drosophila apoptosome
7 c3iz8C_
100.0
17
PDB header: apoptosisChain: C: PDB Molecule: apaf-1 related killer dark;PDBTitle: structure of the drosophila apoptosome
8 c3iz8H_
100.0
17
PDB header: apoptosisChain: H: PDB Molecule: apaf-1 related killer dark;PDBTitle: structure of the drosophila apoptosome
9 c3iz8G_
100.0
17
PDB header: apoptosisChain: G: PDB Molecule: apaf-1 related killer dark;PDBTitle: structure of the drosophila apoptosome
10 c1vt4I_
100.0
17
PDB header: apoptosisChain: I: PDB Molecule: apaf-1 related killer dark;PDBTitle: structure of the drosophila apoptosome
11 c1vt4O_
100.0
17
PDB header: apoptosisChain: O: PDB Molecule: apaf-1 related killer dark;PDBTitle: structure of the drosophila apoptosome
12 c1vt4L_
100.0
17
PDB header: apoptosisChain: L: PDB Molecule: apaf-1 related killer dark;PDBTitle: structure of the drosophila apoptosome
13 c3iz8D_
100.0
17
PDB header: apoptosisChain: D: PDB Molecule: apaf-1 related killer dark;PDBTitle: structure of the drosophila apoptosome
14 c1vt4J_
100.0
17
PDB header: apoptosisChain: J: PDB Molecule: apaf-1 related killer dark;PDBTitle: structure of the drosophila apoptosome
15 c1vt4M_
100.0
17
PDB header: apoptosisChain: M: PDB Molecule: apaf-1 related killer dark;PDBTitle: structure of the drosophila apoptosome
16 c3iz8A_
100.0
17
PDB header: apoptosisChain: A: PDB Molecule: apaf-1 related killer dark;PDBTitle: structure of the drosophila apoptosome
17 c1vt4P_
100.0
17
PDB header: apoptosisChain: P: PDB Molecule: apaf-1 related killer dark;PDBTitle: structure of the drosophila apoptosome
18 c3iz8E_
100.0
17
PDB header: apoptosisChain: E: PDB Molecule: apaf-1 related killer dark;PDBTitle: structure of the drosophila apoptosome
19 c3iz8B_
100.0
17
PDB header: apoptosisChain: B: PDB Molecule: apaf-1 related killer dark;PDBTitle: structure of the drosophila apoptosome
20 c1vt4N_
100.0
17
PDB header: apoptosisChain: N: PDB Molecule: apaf-1 related killer dark;PDBTitle: structure of the drosophila apoptosome
21 c1vt4K_
not modelled
100.0
17
PDB header: apoptosisChain: K: PDB Molecule: apaf-1 related killer dark;PDBTitle: structure of the drosophila apoptosome
22 c2qenA_
not modelled
99.9
14
PDB header: unknown functionChain: A: PDB Molecule: walker-type atpase;PDBTitle: the walker-type atpase paby2304 of pyrococcus abyssi
23 c6j5tC_
not modelled
99.9
15
PDB header: plant proteinChain: C: PDB Molecule: disease resistance rpp13-like protein 4;PDBTitle: reconstitution and structure of a plant nlr resistosome conferring2 immunity
24 c2fnaA_
not modelled
99.9
11
PDB header: atp-binding proteinChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of an archaeal aaa+ atpase (sso1545) from sulfolobus2 solfataricus p2 at 2.00 a resolution
25 c4xgcE_
not modelled
99.8
15
PDB header: dna binding proteinChain: E: PDB Molecule: origin recognition complex subunit 5;PDBTitle: crystal structure of the eukaryotic origin recognition complex
26 c2qbyB_
not modelled
99.8
15
PDB header: replication/dnaChain: B: PDB Molecule: cell division control protein 6 homolog 3;PDBTitle: crystal structure of a heterodimer of cdc6/orc1 initiators2 bound to origin dna (from s. solfataricus)
27 c1fnnB_
not modelled
99.8
14
PDB header: cell cycleChain: B: PDB Molecule: cell division control protein 6;PDBTitle: crystal structure of cdc6p from pyrobaculum aerophilum
28 c6s2pN_
not modelled
99.8
20
PDB header: plant proteinChain: N: PDB Molecule: nrc1;PDBTitle: structure of the nb-arc domain from the tomato immune receptor nrc1
29 c2qbyA_
not modelled
99.8
12
PDB header: replication/dnaChain: A: PDB Molecule: cell division control protein 6 homolog 1;PDBTitle: crystal structure of a heterodimer of cdc6/orc1 initiators2 bound to origin dna (from s. solfataricus)
30 c5ujmE_
not modelled
99.7
12
PDB header: replicationChain: E: PDB Molecule: origin recognition complex subunit 5;PDBTitle: structure of the active form of human origin recognition complex and2 its atpase motor module
31 c2v1uA_
not modelled
99.7
13
PDB header: replicationChain: A: PDB Molecule: cell division control protein 6 homolog;PDBTitle: structure of the aeropyrum pernix orc1 protein in complex2 with dna
32 c1w5sB_
not modelled
99.6
15
PDB header: replicationChain: B: PDB Molecule: origin recognition complex subunit 2 orc2;PDBTitle: structure of the aeropyrum pernix orc2 protein (adp form)
33 c5udb9_
not modelled
99.6
13
PDB header: replicationChain: 9: PDB Molecule: cell division control protein 6;PDBTitle: structural basis of mcm2-7 replicative helicase loading by orc-cdc62 and cdt1
34 c5uj7C_
not modelled
99.6
10
PDB header: dna binding proteinChain: C: PDB Molecule: origin recognition complex subunit 4;PDBTitle: structure of the active form of human origin recognition complex2 atpase motor module, complex subunits 1, 4, 5
35 c5udbA_
not modelled
99.6
8
PDB header: replicationChain: A: PDB Molecule: origin recognition complex subunit 1;PDBTitle: structural basis of mcm2-7 replicative helicase loading by orc-cdc62 and cdt1
36 d2fnaa2
not modelled
99.5
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
37 c5uj7B_
not modelled
99.5
15
PDB header: dna binding proteinChain: B: PDB Molecule: origin recognition complex subunit 1;PDBTitle: structure of the active form of human origin recognition complex2 atpase motor module, complex subunits 1, 4, 5
38 c4xgcD_
not modelled
99.5
23
PDB header: dna binding proteinChain: D: PDB Molecule: origin recognition complex subunit 4;PDBTitle: crystal structure of the eukaryotic origin recognition complex
39 c4kxfP_
not modelled
99.4
17
PDB header: immune systemChain: P: PDB Molecule: nlr family card domain-containing protein 4;PDBTitle: crystal structure of nlrc4 reveals its autoinhibition mechanism
40 c4kxfF_
not modelled
99.4
17
PDB header: immune systemChain: F: PDB Molecule: nlr family card domain-containing protein 4;PDBTitle: crystal structure of nlrc4 reveals its autoinhibition mechanism
41 d1fnna2
not modelled
99.4
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
42 c4xgcA_
not modelled
99.4
15
PDB header: dna binding proteinChain: A: PDB Molecule: origin recognition complex subunit 1;PDBTitle: crystal structure of the eukaryotic origin recognition complex
43 c6npyA_
not modelled
99.2
19
PDB header: immune systemChain: A: PDB Molecule: nacht, lrr and pyd domains-containing protein 3;PDBTitle: cryo-em structure of nlrp3 bound to nek7
44 d1w5sa2
not modelled
99.2
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
45 c5yudA_
not modelled
99.2
13
PDB header: immune systemChain: A: PDB Molecule: baculoviral iap repeat-containing protein 1e;PDBTitle: flagellin derivative in complex with the nlr protein naip5
46 c6b5bA_
not modelled
99.2
15
PDB header: immune systemChain: A: PDB Molecule: baculoviral iap repeat-containing protein 1e;PDBTitle: cryo-em structure of the naip5-nlrc4-flagellin inflammasome
47 c6blbA_
not modelled
99.1
18
PDB header: hydrolaseChain: A: PDB Molecule: holliday junction atp-dependent dna helicase ruvb;PDBTitle: 1.88 angstrom resolution crystal structure holliday junction atp-2 dependent dna helicase (ruvb) from pseudomonas aeruginosa in complex3 with adp
48 c2chgB_
not modelled
99.1
14
PDB header: dna-binding proteinChain: B: PDB Molecule: replication factor c small subunit;PDBTitle: replication factor c domains 1 and 2
49 c1sxjC_
not modelled
99.1
16
PDB header: replicationChain: C: PDB Molecule: activator 1 40 kda subunit;PDBTitle: crystal structure of the eukaryotic clamp loader (replication factor2 c, rfc) bound to the dna sliding clamp (proliferating cell nuclear3 antigen, pcna)
50 d1sxjb2
not modelled
99.1
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
51 c5zr1A_
not modelled
99.0
10
PDB header: dna binding protein/dnaChain: A: PDB Molecule: origin recognition complex subunit 1;PDBTitle: saccharomyces cerevisiae origin recognition complex bound to a 72-bp2 origin dna containing acs and b1 element
52 c3pfiB_
not modelled
99.0
16
PDB header: hydrolaseChain: B: PDB Molecule: holliday junction atp-dependent dna helicase ruvb;PDBTitle: 2.7 angstrom resolution crystal structure of a probable holliday2 junction dna helicase (ruvb) from campylobacter jejuni subsp. jejuni3 nctc 11168 in complex with adenosine-5'-diphosphate
53 c1hqcB_
not modelled
99.0
16
PDB header: hydrolaseChain: B: PDB Molecule: ruvb;PDBTitle: structure of ruvb from thermus thermophilus hb8
54 c2chvE_
not modelled
99.0
13
PDB header: dna-binding proteinChain: E: PDB Molecule: replication factor c small subunit;PDBTitle: replication factor c adpnp complex
55 c5kneA_
not modelled
99.0
22
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein 104;PDBTitle: cryoem reconstruction of hsp104 hexamer
56 c1sxjD_
not modelled
99.0
11
PDB header: replicationChain: D: PDB Molecule: activator 1 41 kda subunit;PDBTitle: crystal structure of the eukaryotic clamp loader (replication factor2 c, rfc) bound to the dna sliding clamp (proliferating cell nuclear3 antigen, pcna)
57 c1iqpF_
not modelled
99.0
12
PDB header: replicationChain: F: PDB Molecule: rfcs;PDBTitle: crystal structure of the clamp loader small subunit from2 pyrococcus furiosus
58 c1sxjB_
not modelled
98.9
11
PDB header: replicationChain: B: PDB Molecule: activator 1 37 kda subunit;PDBTitle: crystal structure of the eukaryotic clamp loader (replication factor2 c, rfc) bound to the dna sliding clamp (proliferating cell nuclear3 antigen, pcna)
59 d1sxjd2
not modelled
98.9
11
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
60 d1sxjc2
not modelled
98.9
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
61 c5udbE_
not modelled
98.9
17
PDB header: replicationChain: E: PDB Molecule: origin recognition complex subunit 5;PDBTitle: structural basis of mcm2-7 replicative helicase loading by orc-cdc62 and cdt1
62 c3te6A_
not modelled
98.9
11
PDB header: gene regulationChain: A: PDB Molecule: regulatory protein sir3;PDBTitle: crystal structure of the s. cerevisiae sir3 aaa+ domain
63 c5d4wB_
not modelled
98.9
21
PDB header: chaperoneChain: B: PDB Molecule: putative heat shock protein;PDBTitle: crystal structure of hsp104
64 d1jbka_
not modelled
98.9
23
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
65 c5kneF_
not modelled
98.9
25
PDB header: chaperoneChain: F: PDB Molecule: heat shock protein 104;PDBTitle: cryoem reconstruction of hsp104 hexamer
66 d1r6bx2
not modelled
98.9
25
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
67 d1iqpa2
not modelled
98.9
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
68 c3pvsA_
not modelled
98.9
16
PDB header: recombinationChain: A: PDB Molecule: replication-associated recombination protein a;PDBTitle: structure and biochemical activities of escherichia coli mgsa
69 c6e111_
not modelled
98.9
18
PDB header: protein transportChain: 1: PDB Molecule: heat shock protein 101;PDBTitle: ptex core complex in the resetting (compact) state
70 d1qvra2
not modelled
98.9
23
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
71 c5kneD_
not modelled
98.8
20
PDB header: chaperoneChain: D: PDB Molecule: heat shock protein 104;PDBTitle: cryoem reconstruction of hsp104 hexamer
72 c6djuA_
not modelled
98.8
27
PDB header: chaperoneChain: A: PDB Molecule: chaperone protein clpb;PDBTitle: mtb clpb in complex with atpgammas and casein, conformer 1
73 c6em8H_
not modelled
98.8
29
PDB header: chaperoneChain: H: PDB Molecule: atp-dependent clp protease atp-binding subunit clpc;PDBTitle: s.aureus clpc resting state, c2 symmetrised
74 c4ciuA_
not modelled
98.8
26
PDB header: chaperoneChain: A: PDB Molecule: chaperone protein clpb;PDBTitle: crystal structure of e. coli clpb
75 c1in8A_
not modelled
98.8
24
PDB header: dna binding proteinChain: A: PDB Molecule: holliday junction dna helicase ruvb;PDBTitle: thermotoga maritima ruvb t158v
76 c6djvE_
not modelled
98.8
27
PDB header: chaperoneChain: E: PDB Molecule: chaperone protein clpb;PDBTitle: mtb clpb in complex with atpgammas and casein, conformer 2
77 c2p65A_
not modelled
98.7
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pf08_0063;PDBTitle: crystal structure of the first nucleotide binding domain of2 chaperone clpb1, putative, (pv089580) from plasmodium vivax
78 c1sxjA_
not modelled
98.7
15
PDB header: replicationChain: A: PDB Molecule: activator 1 95 kda subunit;PDBTitle: crystal structure of the eukaryotic clamp loader (replication factor2 c, rfc) bound to the dna sliding clamp (proliferating cell nuclear3 antigen, pcna)
79 c3bosA_
not modelled
98.7
17
PDB header: hydrolase regulator,dna binding proteinChain: A: PDB Molecule: putative dna replication factor;PDBTitle: crystal structure of a putative dna replication regulator hda2 (sama_1916) from shewanella amazonensis sb2b at 1.75 a resolution
80 d1njfa_
not modelled
98.7
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
81 c5udbD_
not modelled
98.7
11
PDB header: replicationChain: D: PDB Molecule: origin recognition complex subunit 4;PDBTitle: structural basis of mcm2-7 replicative helicase loading by orc-cdc62 and cdt1
82 c6em8F_
not modelled
98.6
25
PDB header: chaperoneChain: F: PDB Molecule: atp-dependent clp protease atp-binding subunit clpc;PDBTitle: s.aureus clpc resting state, c2 symmetrised
83 c1xxhB_
not modelled
98.6
16
PDB header: transferaseChain: B: PDB Molecule: dna polymerase iii subunit gamma;PDBTitle: atpgs bound e. coli clamp loader complex
84 c1r6bX_
not modelled
98.6
18
PDB header: hydrolaseChain: X: PDB Molecule: clpa protein;PDBTitle: high resolution crystal structure of clpa
85 c3pxgA_
not modelled
98.6
32
PDB header: protein bindingChain: A: PDB Molecule: negative regulator of genetic competence clpc/mecb;PDBTitle: structure of meca121 and clpc1-485 complex
86 c1qvrB_
not modelled
98.6
27
PDB header: chaperoneChain: B: PDB Molecule: clpb protein;PDBTitle: crystal structure analysis of clpb
87 c3pxiB_
not modelled
98.6
33
PDB header: protein bindingChain: B: PDB Molecule: negative regulator of genetic competence clpc/mecb;PDBTitle: structure of meca108:clpc
88 d1sxja2
not modelled
98.5
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
89 c5kzfJ_
not modelled
98.5
13
PDB header: hydrolaseChain: J: PDB Molecule: proteasome-associated atpase;PDBTitle: crystal structure of near full-length hexameric mycobacterium2 tuberculosis proteasomal atpase mpa in apo form
90 c6em8E_
not modelled
98.5
19
PDB header: chaperoneChain: E: PDB Molecule: atp-dependent clp protease atp-binding subunit clpc;PDBTitle: s.aureus clpc resting state, c2 symmetrised
91 c5vy9C_
not modelled
98.5
25
PDB header: chaperoneChain: C: PDB Molecule: heat shock protein 104;PDBTitle: s. cerevisiae hsp104:casein complex, middle domain conformation
92 d1ny5a2
not modelled
98.5
15
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
93 c6matE_
not modelled
98.5
23
PDB header: ribosomal proteinChain: E: PDB Molecule: rix7 mutant;PDBTitle: cryo-em structure of the essential ribosome assembly aaa-atpase rix7
94 d1ixsb2
not modelled
98.5
27
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
95 c1ojlF_
not modelled
98.5
14
PDB header: response regulatorChain: F: PDB Molecule: transcriptional regulatory protein zrar;PDBTitle: crystal structure of a sigma54-activator suggests the mechanism for2 the conformational switch necessary for sigma54 binding
96 d1sxje2
not modelled
98.4
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
97 c5x06G_
not modelled
98.4
18
PDB header: replicationChain: G: PDB Molecule: dnaa regulatory inactivator hda;PDBTitle: dna replication regulation protein
98 c6azyA_
not modelled
98.4
30
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein hsp104;PDBTitle: crystal structure of hsp104 r328m/r757m mutant from calcarisporiella2 thermophila
99 c3uk6H_
not modelled
98.4
25
PDB header: hydrolaseChain: H: PDB Molecule: ruvb-like 2;PDBTitle: crystal structure of the tip48 (tip49b) hexamer
100 c3hteC_
not modelled
98.4
22
PDB header: motor proteinChain: C: PDB Molecule: atp-dependent clp protease atp-binding subunit clpx;PDBTitle: crystal structure of nucleotide-free hexameric clpx
101 c1sxjE_
not modelled
98.4
25
PDB header: replicationChain: E: PDB Molecule: activator 1 40 kda subunit;PDBTitle: crystal structure of the eukaryotic clamp loader (replication factor2 c, rfc) bound to the dna sliding clamp (proliferating cell nuclear3 antigen, pcna)
102 d1in4a2
not modelled
98.4
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain
103 c2c9oC_
not modelled
98.4
28
PDB header: hydrolaseChain: C: PDB Molecule: ruvb-like 1;PDBTitle: 3d structure of the human ruvb-like helicase ruvbl1
104 c5ep4A_
not modelled
98.3
20
PDB header: transcriptionChain: A: PDB Molecule: putative repressor protein luxo;PDBTitle: structure, regulation, and inhibition of the quorum-sensing signal2 integrator luxo
105 c2r44A_
not modelled
98.3
13
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative atpase (chu_0153) from cytophaga2 hutchinsonii atcc 33406 at 2.00 a resolution
106 c4z8xC_
not modelled
98.3
15
PDB header: hydrolaseChain: C: PDB Molecule: atp-dependent zinc metalloprotease ftsh;PDBTitle: truncated ftsh from a. aeolicus
107 c2ce7B_
not modelled
98.3
16
PDB header: cell division proteinChain: B: PDB Molecule: cell division protein ftsh;PDBTitle: edta treated
108 c4d2qC_
not modelled
98.3
25
PDB header: chaperoneChain: C: PDB Molecule: clpb;PDBTitle: negative-stain electron microscopy of e. coli clpb mutant e432a (bap2 form bound to clpp)
109 c2z4rB_
not modelled
98.3
15
PDB header: dna binding proteinChain: B: PDB Molecule: chromosomal replication initiator protein dnaa;PDBTitle: crystal structure of domain iii from the thermotoga2 maritima replication initiation protein dnaa
110 c4lcbA_
not modelled
98.3
15
PDB header: protein transportChain: A: PDB Molecule: cell division protein cdvc, vps4;PDBTitle: structure of vps4 homolog from acidianus hospitalis
111 c3d8bB_
not modelled
98.3
15
PDB header: hydrolaseChain: B: PDB Molecule: fidgetin-like protein 1;PDBTitle: crystal structure of human fidgetin-like protein 1 in complex with adp
112 c5exsA_
not modelled
98.3
16
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator fleq;PDBTitle: aaa+ atpase fleq from pseudomonas aeruginosa bound to atp-gamma-s
113 c1xwiA_
not modelled
98.3
15
PDB header: protein transportChain: A: PDB Molecule: skd1 protein;PDBTitle: crystal structure of vps4b
114 c3u5zM_
not modelled
98.3
13
PDB header: dna binding protein/dnaChain: M: PDB Molecule: dna polymerase accessory protein 44;PDBTitle: structure of t4 bacteriophage clamp loader bound to the t4 clamp,2 primer-template dna, and atp analog
115 c2dhrC_
not modelled
98.3
14
PDB header: hydrolaseChain: C: PDB Molecule: ftsh;PDBTitle: whole cytosolic region of atp-dependent metalloprotease2 ftsh (g399l)
116 c2kjqA_
not modelled
98.3
21
PDB header: replicationChain: A: PDB Molecule: dnaa-related protein;PDBTitle: solution structure of protein nmb1076 from neisseria meningitidis.2 northeast structural genomics consortium target mr101b.
117 c6az0A_
not modelled
98.3
41
PDB header: hydrolaseChain: A: PDB Molecule: mitochondrial inner membrane i-aaa protease supercomplexPDBTitle: mitochondrial atpase protease yme1
118 c2qz4A_
not modelled
98.2
17
PDB header: hydrolaseChain: A: PDB Molecule: paraplegin;PDBTitle: human paraplegin, aaa domain in complex with adp
119 c3nbxX_
not modelled
98.2
16
PDB header: hydrolaseChain: X: PDB Molecule: atpase rava;PDBTitle: crystal structure of e. coli rava (regulatory atpase variant a) in2 complex with adp
120 c3eihB_
not modelled
98.2
18
PDB header: protein transportChain: B: PDB Molecule: vacuolar protein sorting-associated protein 4;PDBTitle: crystal structure of s.cerevisiae vps4 in the presence of atpgammas