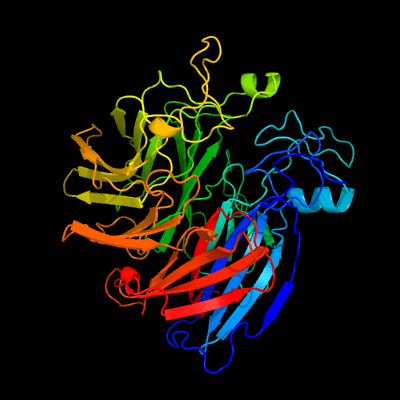
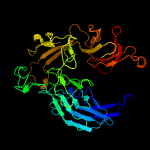
1 c5j54A_
100.0
27
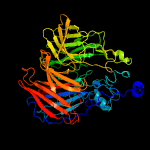
PDB header: oxidoreductaseChain: A: PDB Molecule: carotenoid oxygenase;PDBTitle: the structure and mechanism of nov1, a resveratrol-cleaving2 dioxygenase
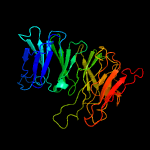
2 c5v2dA_
100.0
28
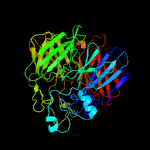
PDB header: oxidoreductaseChain: A: PDB Molecule: dioxygenase;PDBTitle: crystal structure of pseudomonas brassicacearum lignostilbene2 dioxygenase
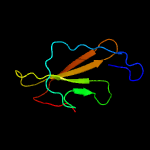
3 c5u90A_
100.0
26
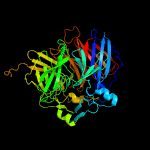
PDB header: oxidoreductaseChain: A: PDB Molecule: carotenoid oxygenase 1;PDBTitle: crystal structure of co-cao1 in complex with resveratrol
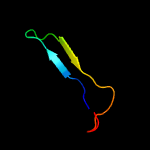
4 c3npeA_
100.0
28
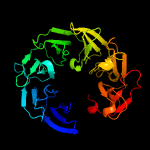
PDB header: oxidoreductaseChain: A: PDB Molecule: 9-cis-epoxycarotenoid dioxygenase 1, chloroplastic;PDBTitle: structure of vp14 in complex with oxygen
5 c2biwC_
100.0
25
PDB header: oxidoreductaseChain: C: PDB Molecule: apocarotenoid-cleaving oxygenase;PDBTitle: crystal structure of apocarotenoid cleavage oxygenase from2 synechocystis, native enzyme
6 c3fsnA_
100.0
18
PDB header: isomeraseChain: A: PDB Molecule: retinal pigment epithelium-specific 65 kda protein;PDBTitle: crystal structure of rpe65 at 2.14 angstrom resolution
7 c6rteB_
83.4
16
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome c;PDBTitle: dihydro-heme d1 dehydrogenase nirn in complex with dhe
8 c4cvcA_
77.4
10
PDB header: oxidoreductaseChain: A: PDB Molecule: alcohol dehydrogenase;PDBTitle: crystal structure of quinone-dependent alcohol dehydrogenase from2 pseudogluconobacter saccharoketogenenes with zinc in the active site
9 c1nnoA_
57.4
14
PDB header: oxidoreductaseChain: A: PDB Molecule: nitrite reductase;PDBTitle: conformational changes occurring upon no binding in nitrite reductase2 from pseudomonas aeruginosa
10 d1mdah_
39.4
11
Fold: 7-bladed beta-propellerSuperfamily: YVTN repeat-like/Quinoprotein amine dehydrogenaseFamily: Methylamine dehydrogenase, H-chain
11 c5dilA_
31.2
32
PDB header: rna-binding protein, viral proteinChain: A: PDB Molecule: non-structural protein 1;PDBTitle: crystal structure of the effector domain of the ns1 protein from2 influenza virus b
12 d2hu7a1
26.9
15
Fold: 7-bladed beta-propellerSuperfamily: Peptidase/esterase 'gauge' domainFamily: Acylamino-acid-releasing enzyme, N-terminal donain
13 c3qc2A_
26.4
28
PDB header: hydrolaseChain: A: PDB Molecule: glycosyl hydrolase;PDBTitle: crystal structure of a glycosyl hydrolase (bacova_03624) from2 bacteroides ovatus at 2.30 a resolution
14 c5aycA_
25.2
21
PDB header: transferaseChain: A: PDB Molecule: 4-o-beta-d-mannosyl-d-glucose phosphorylase;PDBTitle: crystal structure of ruminococcus albus 4-o-beta-d-mannosyl-d-glucose2 phosphorylase (ramp1) in complexes with sulfate and 4-o-beta-d-3 mannosyl-d-glucose
15 c5f30B_
21.4
18
PDB header: oxidoreductaseChain: B: PDB Molecule: thiocyanate dehydrogenase;PDBTitle: thiocyanate dehydrogenase from thioalkalivibrio paradoxus
16 d1zaka2
21.4
32
Fold: Rubredoxin-likeSuperfamily: Microbial and mitochondrial ADK, insert "zinc finger" domainFamily: Microbial and mitochondrial ADK, insert "zinc finger" domain
17 c5c2wD_
21.2
13
PDB header: oxidoreductaseChain: D: PDB Molecule: hydrazine synthase alpha subunit;PDBTitle: kuenenia stuttgartiensis hydrazine synthase pressurized with 20 bar2 xenon
18 c3watA_
17.2
17
PDB header: transferaseChain: A: PDB Molecule: 4-o-beta-d-mannosyl-d-glucose phosphorylase;PDBTitle: crystal structure of 4-o-beta-d-mannosyl-d-glucose phosphorylase mgp2 complexed with man+glc
19 c3metB_
15.8
18
PDB header: transcriptionChain: B: PDB Molecule: saga-associated factor 29 homolog;PDBTitle: crystal structure of sgf29 in complex with h3k4me2
20 c3elqA_
14.5
11
PDB header: transferaseChain: A: PDB Molecule: arylsulfate sulfotransferase;PDBTitle: crystal structure of a bacterial arylsulfate2 sulfotransferase
21 d1jmxb_
not modelled
14.4
18
Fold: 7-bladed beta-propellerSuperfamily: YVTN repeat-like/Quinoprotein amine dehydrogenaseFamily: Quinohemoprotein amine dehydrogenase B chain
22 c4ozuA_
not modelled
12.8
14
PDB header: structural proteinChain: A: PDB Molecule: coronin;PDBTitle: crystal structure of wd40 domain from toxoplasma gondii coronin
23 c6cmkB_
not modelled
12.8
17
PDB header: metal binding proteinChain: B: PDB Molecule: aztd protein;PDBTitle: crystal structure of citrobacter koseri aztd
24 d1m7ja2
not modelled
12.6
35
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: D-aminoacylase
25 c2vb6A_
not modelled
12.0
20
PDB header: motor proteinChain: A: PDB Molecule: myosin vi;PDBTitle: myosin vi (md-insert2-cam, delta insert1) post-rigor state (2 crystal form 2)
26 d1mspa_
not modelled
11.5
36
Fold: Immunoglobulin-like beta-sandwichSuperfamily: PapD-likeFamily: MSP-like
27 c1gq1B_
not modelled
11.5
12
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome cd1 nitrite reductase;PDBTitle: cytochrome cd1 nitrite reductase, y25s mutant, oxidised form
28 c3knuD_
not modelled
11.3
25
PDB header: transferaseChain: D: PDB Molecule: trna (guanine-n(1)-)-methyltransferase;PDBTitle: crystal structure of trna (guanine-n1)-methyltransferase from2 anaplasma phagocytophilum
29 c5i5iA_
not modelled
11.2
8
PDB header: oxidoreductaseChain: A: PDB Molecule: nitrous-oxide reductase;PDBTitle: shewanella denitrificans nitrous oxide reductase, app form
30 d1jofa_
not modelled
11.2
8
Fold: 7-bladed beta-propellerSuperfamily: 3-carboxy-cis,cis-mucoante lactonizing enzymeFamily: 3-carboxy-cis,cis-mucoante lactonizing enzyme
31 c1kv9A_
not modelled
10.8
13
PDB header: oxidoreductaseChain: A: PDB Molecule: type ii quinohemoprotein alcohol dehydrogenase;PDBTitle: structure at 1.9 a resolution of a quinohemoprotein alcohol2 dehydrogenase from pseudomonas putida hk5
32 d1k91a_
not modelled
10.5
13
Fold: P-domain of calnexin/calreticulinSuperfamily: P-domain of calnexin/calreticulinFamily: P-domain of calnexin/calreticulin
33 c5c2mA_
not modelled
10.2
35
PDB header: structural proteinChain: A: PDB Molecule: predicted protein;PDBTitle: the de novo evolutionary emergence of a symmetrical protein is shaped2 by folding constraints
34 d2oz4a1
not modelled
9.7
27
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: C2 set domains
35 c5uqdA_
not modelled
9.4
22
PDB header: oxidoreductaseChain: A: PDB Molecule: dumpy: shorter than wild-type;PDBTitle: dpy-21 in complex with fe(ii) and alpha-ketoglutarate
36 d2cu6a1
not modelled
8.6
24
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: PaaD-like
37 d2noha2
not modelled
7.8
8
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: DNA repair glycosylase, N-terminal domain
38 c2bkiA_
not modelled
7.7
20
PDB header: motor protein/metal-binding proteinChain: A: PDB Molecule: unconventional myosin;PDBTitle: myosin vi nucleotide-free (mdinsert2-iq) crystal structure
39 c6cslA_
not modelled
7.6
25
PDB header: metal binding proteinChain: A: PDB Molecule: histidine triad protein d;PDBTitle: pneumococcal phtd protein 269-339 fragment with bound zn(ii)
40 c1bb9A_
not modelled
7.6
18
PDB header: transferaseChain: A: PDB Molecule: amphiphysin 2;PDBTitle: crystal structure of the sh3 domain from rat amphiphysin 2
41 d1bb9a_
not modelled
7.6
18
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain
42 c3e7hA_
not modelled
7.4
53
PDB header: transferaseChain: A: PDB Molecule: dna-directed rna polymerase subunit beta;PDBTitle: the crystal structure of the beta subunit of the dna-directed rna2 polymerase from vibrio cholerae o1 biovar eltor
43 c1mv3A_
not modelled
7.2
16
PDB header: endocytosis/exocytosisChain: A: PDB Molecule: myc box dependent interacting protein 1;PDBTitle: nmr structure of the tumor suppressor bin1: alternative2 splicing in melanoma and interaction with c-myc
44 d2fe0a1
not modelled
7.2
12
Fold: Smp-1-likeSuperfamily: Smp-1-likeFamily: Smp-1-like
45 d1xpna_
not modelled
7.1
12
Fold: Prealbumin-likeSuperfamily: Hypothetical protein PA1324Family: Hypothetical protein PA1324
46 d1njha_
not modelled
7.1
26
Fold: Hypothetical protein YojFSuperfamily: Hypothetical protein YojFFamily: Hypothetical protein YojF
47 c6cfwI_
not modelled
7.0
15
PDB header: membrane proteinChain: I: PDB Molecule: mbh subunit;PDBTitle: cryoem structure of a respiratory membrane-bound hydrogenase
48 c2fu4B_
not modelled
6.9
27
PDB header: dna binding proteinChain: B: PDB Molecule: ferric uptake regulation protein;PDBTitle: crystal structure of the dna binding domain of e.coli fur (ferric2 uptake regulator)
49 c5oxzB_
not modelled
6.8
22
PDB header: splicingChain: B: PDB Molecule: neq528;PDBTitle: crystal structure of neqn/neqc complex from nanoarcheaum equitans at2 1.2a
50 c4c95C_
not modelled
6.8
11
PDB header: dna replicationChain: C: PDB Molecule: dna polymerase alpha-binding protein;PDBTitle: crystal structure of the carboxy-terminal domain of yeast2 ctf4 bound to sld5
51 c2bkhA_
not modelled
6.7
20
PDB header: motor protein/metal-binding proteinChain: A: PDB Molecule: unconventional myosin;PDBTitle: myosin vi nucleotide-free (mdinsert2) crystal structure
52 c6g72A_
not modelled
6.5
18
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh-ubiquinone oxidoreductase chain 3;PDBTitle: mouse mitochondrial complex i in the deactive state
53 d2cs7a1
not modelled
6.5
25
Fold: IL8-likeSuperfamily: PhtA domain-likeFamily: PhtA domain-like
54 c2ghsA_
not modelled
6.4
13
PDB header: calcium-binding proteinChain: A: PDB Molecule: agr_c_1268p;PDBTitle: crystal structure of a calcium-binding protein, regucalcin2 (agr_c_1268) from agrobacterium tumefaciens str. c58 at 1.55 a3 resolution
55 d2ghsa1
not modelled
6.4
13
Fold: 6-bladed beta-propellerSuperfamily: Calcium-dependent phosphotriesteraseFamily: SGL-like
56 c5tbyA_
not modelled
6.3
3
PDB header: contractile proteinChain: A: PDB Molecule: myosin-7;PDBTitle: human beta cardiac heavy meromyosin interacting-heads motif obtained2 by homology modeling (using swiss-model) of human sequence from3 aphonopelma homology model (pdb-3jbh), rigidly fitted to human beta-4 cardiac negatively stained thick filament 3d-reconstruction (emd-5 2240)
57 d1bf2a1
not modelled
6.2
10
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes
58 c4fc9B_
not modelled
6.0
25
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: structure of the c-terminal domain of the type iii effector xcv32202 (xopl)
59 c3q54A_
not modelled
6.0
16
PDB header: lipid binding proteinChain: A: PDB Molecule: outer membrane assembly lipoprotein yfgl;PDBTitle: crystal structure of escherichia coli bamb
60 d1xv2a_
not modelled
5.9
19
Fold: AF0104/ALDC/Ptd012-likeSuperfamily: AF0104/ALDC/Ptd012-likeFamily: Alpha-acetolactate decarboxylase-like
61 c4wndB_
not modelled
5.9
13
PDB header: signaling protein/protein bindingChain: B: PDB Molecule: ferm and pdz domain-containing protein 4;PDBTitle: crystal structure of the tpr domain of lgn in complex with2 frmpd4/preso1 at 1.5 angstrom resolution
62 c5v1dA_
not modelled
5.9
14
PDB header: transferase/substrateChain: A: PDB Molecule: eif2ak3 protein;PDBTitle: complex structure of the bovine perk luminal domain and its substrate2 peptide
63 c1yr2A_
not modelled
5.8
11
PDB header: hydrolaseChain: A: PDB Molecule: prolyl oligopeptidase;PDBTitle: structural and mechanistic analysis of two prolyl endopeptidases: role2 of inter-domain dynamics in catalysis and specificity
64 c4c8hC_
not modelled
5.8
12
PDB header: dna replicationChain: C: PDB Molecule: ctf4;PDBTitle: crystal structure of the c-terminal region of yeast ctf4,2 selenomethionine protein.
65 c5yhoA_
not modelled
5.7
28
PDB header: metal binding proteinChain: A: PDB Molecule: alpha-acetolactate decarboxylase;PDBTitle: crystal structure of acetolactate decarboxylase from enterobacter2 cloacae
66 c4wneB_
not modelled
5.7
13
PDB header: signaling protein/protein bindingChain: B: PDB Molecule: peptide from ferm and pdz domain-containing protein 4;PDBTitle: crystal structure of the tpr domain of lgn in complex with2 frmpd4/preso1 at 2.0 angstrom resolution
67 c3mbrX_
not modelled
5.6
13
PDB header: transferaseChain: X: PDB Molecule: glutamine cyclotransferase;PDBTitle: crystal structure of the glutaminyl cyclase from xanthomonas2 campestris
68 d1lmia_
not modelled
5.4
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Antigen MPT63/MPB63 (immunoprotective extracellular protein)Family: Antigen MPT63/MPB63 (immunoprotective extracellular protein)
69 c3grdA_
not modelled
5.4
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized ntf2-superfamily protein;PDBTitle: crystal structure of ntf2-superfamily protein with unknown function2 (np_977240.1) from bacillus cereus atcc 10987 at 1.25 a resolution
70 c4mveB_
not modelled
5.3
16
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of tcur_1030 protein from thermomonospora curvata
71 c5gvbA_
not modelled
5.2
10
PDB header: replication, peptide binding proteinChain: A: PDB Molecule: wd repeat and hmg-box dna-binding protein 1;PDBTitle: sepb domain of human and-1
72 c3mjjD_
not modelled
5.2
16
PDB header: hydrolaseChain: D: PDB Molecule: predicted acetamidase/formamidase;PDBTitle: crystal structure analysis of a recombinant predicted2 acetamidase/formamidase from the thermophile thermoanaerobacter3 tengcongensis
73 d1kv9a2
not modelled
5.2
9
Fold: 8-bladed beta-propellerSuperfamily: Quinoprotein alcohol dehydrogenase-likeFamily: Quinoprotein alcohol dehydrogenase-like
74 c2kyyA_
not modelled
5.1
20
PDB header: hydrolaseChain: A: PDB Molecule: possible atp-dependent dna helicase recg-related protein;PDBTitle: solution nmr structure of the n-terminal domain of putative atp-2 dependent dna helicase recg-related protein from nitrosomonas3 europaea, northeast structural genomics consortium target ner70a
75 c6dk4A_
not modelled
5.1
15
PDB header: metal transportChain: A: PDB Molecule: ferric uptake regulation protein;PDBTitle: crystal structure of campylobacter jejuni peroxide stress regulator
76 d1dq3a4
not modelled
5.0
18
Fold: Homing endonuclease-likeSuperfamily: Homing endonucleasesFamily: Intein endonuclease
77 c5nsaA_
not modelled
5.0
30
PDB header: transport proteinChain: A: PDB Molecule: transcobalamin-2;PDBTitle: beta domain of human transcobalamin bound to co-beta-[2-(2,4-2 difluorophenyl)ethinyl]cobalamin