| 1 |
|
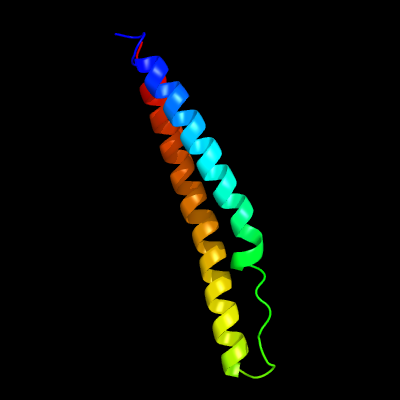
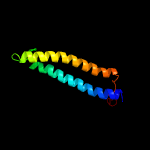
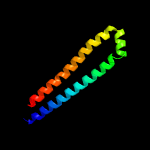
PDB 5xfs chain A
Region: 7 - 84
Aligned: 78
Modelled: 78
Confidence: 100.0%
Identity: 45%
PDB header:protein transport
Chain: A: PDB Molecule:pe family protein pe8;
PDBTitle: crystal structure of pe8-ppe15 in complex with espg5 from m.2 tuberculosis
Phyre2
| 2 |
|
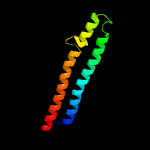
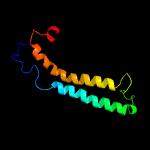
PDB 2g38 chain A domain 1
Region: 8 - 84
Aligned: 77
Modelled: 77
Confidence: 100.0%
Identity: 29%
Fold: Ferritin-like
Superfamily: PE/PPE dimer-like
Family: PE
Phyre2
| 3 |
|
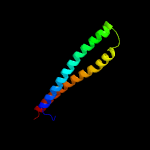
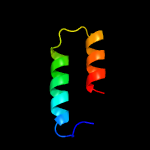
PDB 2g38 chain A
Region: 8 - 84
Aligned: 77
Modelled: 77
Confidence: 100.0%
Identity: 29%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:pe family protein;
PDBTitle: a pe/ppe protein complex from mycobacterium tuberculosis
Phyre2
| 4 |
|
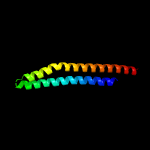
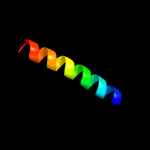
PDB 4wj2 chain A
Region: 17 - 91
Aligned: 75
Modelled: 75
Confidence: 79.8%
Identity: 13%
PDB header:unknown function
Chain: A: PDB Molecule:antigen mtb48;
PDBTitle: mycobacterial protein
Phyre2
| 5 |
|
PDB 3gvm chain A
Region: 1 - 97
Aligned: 93
Modelled: 97
Confidence: 76.0%
Identity: 14%
PDB header:viral protein
Chain: A: PDB Molecule:putative uncharacterized protein sag1039;
PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
Phyre2
| 6 |
|
PDB 1wa8 chain A domain 1
Region: 4 - 97
Aligned: 90
Modelled: 94
Confidence: 67.6%
Identity: 10%
Fold: Ferritin-like
Superfamily: EsxAB dimer-like
Family: ESAT-6 like
Phyre2
| 7 |
|
PDB 4iog chain D
Region: 1 - 90
Aligned: 86
Modelled: 90
Confidence: 60.7%
Identity: 12%
PDB header:unknown function
Chain: D: PDB Molecule:secreted protein esxb;
PDBTitle: the crystal structure of a secreted protein esxb (wild-type, in p212 space group) from bacillus anthracis str. sterne
Phyre2
| 8 |
|
PDB 2vs0 chain B
Region: 4 - 91
Aligned: 84
Modelled: 88
Confidence: 51.8%
Identity: 12%
PDB header:cell invasion
Chain: B: PDB Molecule:virulence factor esxa;
PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
Phyre2
| 9 |
|
PDB 1lgh chain B
Region: 68 - 80
Aligned: 13
Modelled: 13
Confidence: 33.6%
Identity: 31%
Fold: Light-harvesting complex subunits
Superfamily: Light-harvesting complex subunits
Family: Light-harvesting complex subunits
Phyre2
| 10 |
|
PDB 3zbh chain C
Region: 7 - 97
Aligned: 87
Modelled: 91
Confidence: 25.9%
Identity: 10%
PDB header:unknown function
Chain: C: PDB Molecule:esxa;
PDBTitle: geobacillus thermodenitrificans esxa crystal form i
Phyre2
| 11 |
|
PDB 1wrg chain A
Region: 68 - 80
Aligned: 13
Modelled: 13
Confidence: 23.8%
Identity: 23%
PDB header:membrane protein
Chain: A: PDB Molecule:light-harvesting protein b-880, beta chain;
PDBTitle: light-harvesting complex 1 beta subunit from wild-type2 rhodospirillum rubrum
Phyre2
| 12 |
|
PDB 6et5 chain U
Region: 68 - 80
Aligned: 13
Modelled: 13
Confidence: 13.6%
Identity: 38%
PDB header:photosynthesis
Chain: U: PDB Molecule:light-harvesting protein b-1015 gamma chain;
PDBTitle: reaction centre light harvesting complex 1 from blc. virids
Phyre2
| 13 |
|
PDB 4lws chain B
Region: 1 - 87
Aligned: 83
Modelled: 87
Confidence: 9.7%
Identity: 13%
PDB header:unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
Phyre2
| 14 |
|
PDB 2kg7 chain B
Region: 1 - 99
Aligned: 95
Modelled: 99
Confidence: 8.0%
Identity: 11%
PDB header:unknown function
Chain: B: PDB Molecule:esat-6-like protein esxh;
PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
Phyre2
| 15 |
|
PDB 4rgl chain A
Region: 40 - 77
Aligned: 38
Modelled: 37
Confidence: 7.7%
Identity: 18%
PDB header:dna binding protein
Chain: A: PDB Molecule:filamentation induced by camp protein fic;
PDBTitle: crystal structure of a fic family protein (dde_2494) from2 desulfovibrio desulfuricans g20 at 2.70 a resolution
Phyre2
| 16 |
|
PDB 3onj chain A
Region: 54 - 76
Aligned: 23
Modelled: 23
Confidence: 6.9%
Identity: 4%
PDB header:protein transport
Chain: A: PDB Molecule:t-snare vti1;
PDBTitle: crystal structure of yeast vti1p_habc domain
Phyre2
| 17 |
|
PDB 4lws chain A
Region: 6 - 97
Aligned: 88
Modelled: 88
Confidence: 5.5%
Identity: 15%
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
Phyre2