1 c5mjiA_
100.0
38
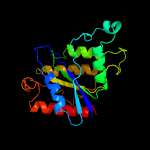
PDB header: flavoproteinChain: A: PDB Molecule: bramp domain protein;PDBTitle: crystal structure of rosb with bound intermediate ohc-rp (8-demethyl-2 8-formylriboflavin-5'-phosphate)
2 d1sqsa_
100.0
16
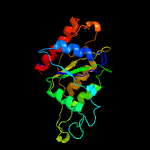
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Hypothetical protein SP1951
3 d1ydga_
100.0
14
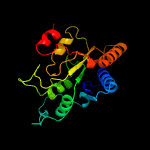
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: WrbA-like
4 c3s2yB_
100.0
15
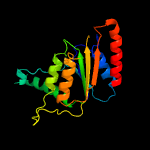
PDB header: oxidoreductaseChain: B: PDB Molecule: chromate reductase;PDBTitle: crystal structure of a chromate/uranium reductase from2 gluconacetobacter hansenii
5 c2zkiH_
100.0
13
PDB header: transcriptionChain: H: PDB Molecule: 199aa long hypothetical trp repressor bindingPDBTitle: crystal structure of hypothetical trp repressor binding2 protein from sul folobus tokodaii (st0872)
6 d1t0ia_
100.0
13
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: NADPH-dependent FMN reductase
7 c3lcmB_
100.0
14
PDB header: oxidoreductaseChain: B: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of smu.1420 from streptococcus mutans ua159
8 c2q62A_
100.0
12
PDB header: flavoproteinChain: A: PDB Molecule: arsh;PDBTitle: crystal structure of arsh from sinorhizobium meliloti
9 c4r81C_
100.0
13
PDB header: oxidoreductaseChain: C: PDB Molecule: nadh dehydrogenase;PDBTitle: nad(p)h:quinone oxidoreductase from methanothermobacter marburgensis
10 c4ptzC_
100.0
16
PDB header: oxidoreductaseChain: C: PDB Molecule: fmn reductase ssue;PDBTitle: crystal structure of the escherichia coli alkanesulfonate fmn2 reductase ssue in fmn-bound form
11 c3svlB_
100.0
15
PDB header: oxidoreductaseChain: B: PDB Molecule: protein yief;PDBTitle: structural basis of the improvement of chrr - a multi-purpose enzyme
12 c3u7rB_
100.0
14
PDB header: oxidoreductaseChain: B: PDB Molecule: nadph-dependent fmn reductase;PDBTitle: ferb - flavoenzyme nad(p)h:(acceptor) oxidoreductase (ferb) from2 paracoccus denitrificans
13 d2qwxa1
100.0
17
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Quinone reductase
14 c3w7aD_
100.0
13
PDB header: oxidoreductaseChain: D: PDB Molecule: fmn-dependent nadh-azoreductase;PDBTitle: crystal structure of azoreductase azrc fin complex with sulfone-2 modified azo dye acid red 88
15 d1rtta_
100.0
12
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: NADPH-dependent FMN reductase
16 d1nni1_
100.0
16
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: NADPH-dependent FMN reductase
17 c2fzvC_
100.0
14
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: putative arsenical resistance protein;PDBTitle: crystal structure of an apo form of a flavin-binding protein from2 shigella flexneri
18 c4c76A_
100.0
10
PDB header: oxidoreductaseChain: A: PDB Molecule: fmn reductase (nadph);PDBTitle: crystal structure of the fmn-reductase msue from pseudomonas putida2 kt2440.
19 c4lafB_
99.9
14
PDB header: oxidoreductaseChain: B: PDB Molecule: nad(p)h dehydrogenase (quinone);PDBTitle: crystal structure of pnpb complex with fmn
20 c6dxpD_
99.9
13
PDB header: oxidoreductaseChain: D: PDB Molecule: fmn-dependent nadh-azoreductase;PDBTitle: the crystal structure of an fmn-dependent nadh-azoreductase from2 klebsiella pneumoniae
21 d1t5ba_
not modelled
99.9
12
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Quinone reductase
22 d2fzva1
not modelled
99.9
15
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: NADPH-dependent FMN reductase
23 c3u7iB_
not modelled
99.9
15
PDB header: oxidoreductaseChain: B: PDB Molecule: fmn-dependent nadh-azoreductase 1;PDBTitle: the crystal structure of fmn-dependent nadh-azoreductase 1 (gbaa0966)2 from bacillus anthracis str. ames ancestor
24 c3b6iB_
not modelled
99.9
13
PDB header: flavoproteinChain: B: PDB Molecule: flavoprotein wrba;PDBTitle: wrba from escherichia coli, native structure
25 c2hpvA_
not modelled
99.9
13
PDB header: oxidoreductaseChain: A: PDB Molecule: fmn-dependent nadh-azoreductase;PDBTitle: crystal structure of fmn-dependent azoreductase from enterococcus2 faecalis
26 c4c0xA_
not modelled
99.9
14
PDB header: oxidoreductaseChain: A: PDB Molecule: fmn-dependent nadh-azoreductase 1;PDBTitle: the crystal strucuture of ppazor in complex with anthraquinone-2-2 sulfonate
27 c3fvwA_
not modelled
99.9
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative nad(p)h-dependent fmn reductase;PDBTitle: crystal structure of the q8dwd8_strmu protein from streptococcus2 mutans. northeast structural genomics consortium target smr99.
28 c2v9cA_
not modelled
99.9
15
PDB header: oxidoreductaseChain: A: PDB Molecule: fmn-dependent nadh-azoreductase 1;PDBTitle: x-ray crystallographic structure of a pseudomonas2 aeruginosa azoreductase in complex with methyl red.
29 c2vzhA_
not modelled
99.9
17
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh-dependent fmn reductase;PDBTitle: structures of nadh:fmn oxidoreductase (emob)-fmn complex
30 c5mp4C_
not modelled
99.9
15
PDB header: oxidoreductaseChain: C: PDB Molecule: protoplast secreted protein 2;PDBTitle: the structure of pst2p from saccharomyces cerevisiae
31 c4gi5B_
not modelled
99.9
18
PDB header: oxidoreductaseChain: B: PDB Molecule: quinone reductase;PDBTitle: crystal structure of a putative quinone reductase from klebsiella2 pneumoniae (target psi-013613)
32 d1d4aa_
not modelled
99.9
13
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Quinone reductase
33 d2z98a1
not modelled
99.9
12
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Quinone reductase
34 d1dxqa_
not modelled
99.9
13
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Quinone reductase
35 d2a5la1
not modelled
99.9
14
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: WrbA-like
36 c3k1yE_
not modelled
99.9
18
PDB header: oxidoreductaseChain: E: PDB Molecule: oxidoreductase;PDBTitle: x-ray structure of oxidoreductase from corynebacterium diphtheriae.2 orthorombic crystal form, northeast structural genomics consortium3 target cdr100d
37 d1qrda_
not modelled
99.9
14
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Quinone reductase
38 c3p0rA_
not modelled
99.9
17
PDB header: oxidoreductaseChain: A: PDB Molecule: azoreductase;PDBTitle: crystal structure of azoreductase from bacillus anthracis str. sterne
39 c5f4bB_
not modelled
99.9
16
PDB header: oxidoreductaseChain: B: PDB Molecule: nad(p)h dehydrogenase (quinone);PDBTitle: structure of b. abortus wrba-related protein a (wrpa)
40 d1rlia_
not modelled
99.9
16
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Hypothetical protein YwqN
41 c5lvaA_
not modelled
99.9
9
PDB header: oxidoreductaseChain: A: PDB Molecule: nad(p)h-fmn oxidoreductase;PDBTitle: crystal structure of thermophilic tryptophan halogenase (th-hal)2 enzyme from streptomycin violaceusniger.
42 c3d7nA_
not modelled
99.9
9
PDB header: electron transportChain: A: PDB Molecule: flavodoxin, wrba-like protein;PDBTitle: the crystal structure of the flavodoxin, wrba-like protein from2 agrobacterium tumefaciens
43 c3rpeA_
not modelled
99.8
23
PDB header: oxidoreductaseChain: A: PDB Molecule: modulator of drug activity b;PDBTitle: 1.1 angstrom crystal structure of putative modulator of drug activity2 (mdab) from yersinia pestis co92.
44 c3f2vA_
not modelled
99.8
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: general stress protein 14;PDBTitle: crystal structure of the general stress protein 14 (tde0354) in2 complex with fmn from treponema denticola, northeast structural3 genomics consortium target tdr58.
45 d2arka1
not modelled
99.8
20
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: WrbA-like
46 d1e5da1
not modelled
99.7
10
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
47 d1ycga1
not modelled
99.7
12
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
48 c6h0cA_
not modelled
99.7
14
PDB header: oxidoreductaseChain: A: PDB Molecule: putative diflavin flavoprotein a 3;PDBTitle: flv1 flavodiiron core from synechocystis sp. pcc6803
49 c2amjD_
not modelled
99.7
26
PDB header: oxidoreductaseChain: D: PDB Molecule: modulator of drug activity b;PDBTitle: crystal structure of modulator of drug activity b from escherichia2 coli o157:h7
50 c1ychD_
not modelled
99.7
12
PDB header: oxidoreductaseChain: D: PDB Molecule: nitric oxide reductase;PDBTitle: x-ray crystal structures of moorella thermoacetica fpra. novel diiron2 site structure and mechanistic insights into a scavenging nitric3 oxide reductase
51 c2ohiB_
not modelled
99.6
13
PDB header: oxidoreductaseChain: B: PDB Molecule: type a flavoprotein fpra;PDBTitle: crystal structure of coenzyme f420h2 oxidase (fpra), a diiron2 flavoprotein, reduced state
52 c2q9uB_
not modelled
99.6
9
PDB header: oxidoreductaseChain: B: PDB Molecule: a-type flavoprotein;PDBTitle: crystal structure of the flavodiiron protein from giardia2 intestinalis
53 c3ha2B_
not modelled
99.6
16
PDB header: oxidoreductaseChain: B: PDB Molecule: nadph-quinone reductase;PDBTitle: crystal structure of protein (nadph-quinone reductase) from2 p.pentosaceus, northeast structural genomics consortium target ptr24a
54 c5widB_
not modelled
99.6
13
PDB header: flavoproteinChain: B: PDB Molecule: flavodoxin;PDBTitle: structure of a flavodoxin from the domain archaea
55 c4d02A_
not modelled
99.6
15
PDB header: electron transportChain: A: PDB Molecule: anaerobic nitric oxide reductase flavorubredoxin;PDBTitle: the crystallographic structure of flavorubredoxin from escherichia2 coli
56 d1vmea1
not modelled
99.5
9
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
57 c1e5dA_
not modelled
99.5
11
PDB header: oxidoreductaseChain: A: PDB Molecule: rubredoxin\:oxygen oxidoreductase;PDBTitle: rubredoxin oxygen:oxidoreductase (roo) from anaerobe desulfovibrio2 gigas
58 c4iciA_
not modelled
99.5
9
PDB header: flavoproteinChain: A: PDB Molecule: putative flavoprotein;PDBTitle: crystal structure of a putative flavoprotein (bacegg_01620) from2 bacteroides eggerthii dsm 20697 at 1.40 a resolution
59 c4j8pA_
not modelled
99.5
8
PDB header: flavoproteinChain: A: PDB Molecule: flavodoxin;PDBTitle: crystal structure of a putative flavoprotein (bacuni_04544) from2 bacteroides uniformis atcc 8492 at 1.50 a resolution
60 c3edoA_
not modelled
99.4
10
PDB header: flavoproteinChain: A: PDB Molecule: putative trp repressor binding protein;PDBTitle: crystal structure of flavoprotein in complex with fmn (yp_193882.1)2 from lactobacillus acidophilus ncfm at 1.20 a resolution
61 c3klbA_
not modelled
99.4
13
PDB header: flavoproteinChain: A: PDB Molecule: putative flavoprotein;PDBTitle: crystal structure of putative flavoprotein in complex with fmn2 (yp_213683.1) from bacteroides fragilis nctc 9343 at 1.75 a3 resolution
62 c3fniA_
not modelled
99.4
17
PDB header: oxidoreductaseChain: A: PDB Molecule: putative diflavin flavoprotein a 3;PDBTitle: crystal structure of a diflavin flavoprotein a3 (all3895) from nostoc2 sp., northeast structural genomics consortium target nsr431a
63 c3hlyA_
not modelled
99.3
20
PDB header: flavoproteinChain: A: PDB Molecule: flavodoxin-like domain;PDBTitle: crystal structure of the flavodoxin-like domain from synechococcus sp2 q5mzp6_synp6 protein. northeast structural genomics consortium target3 snr135d.
64 c3eywA_
not modelled
99.3
15
PDB header: transport proteinChain: A: PDB Molecule: c-terminal domain of glutathione-regulated potassium-effluxPDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
65 c1vmeB_
not modelled
99.3
9
PDB header: electron transportChain: B: PDB Molecule: flavoprotein;PDBTitle: crystal structure of flavoprotein (tm0755) from thermotoga maritima at2 1.80 a resolution
66 c6gaqB_
not modelled
99.0
17
PDB header: electron transportChain: B: PDB Molecule: flavodoxin;PDBTitle: crystal structure of oxidised flavodoxin 2 from bacillus cereus
67 d2fz5a1
not modelled
98.9
17
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
68 c5vegC_
not modelled
98.9
18
PDB header: electron transportChain: C: PDB Molecule: flavodoxin;PDBTitle: structure of a short-chain flavodoxin associated with a non-canonical2 pdu bacterial microcompartment
69 c6fsiA_
not modelled
98.9
18
PDB header: electron transportChain: A: PDB Molecule: flavodoxin;PDBTitle: crystal structure of semiquinone flavodoxin 1 from bacillus cereus2 (1.32 a resolution)
70 d5nula_
not modelled
98.8
16
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
71 c5ljiA_
not modelled
98.6
20
PDB header: oxidoreductaseChain: A: PDB Molecule: flavodoxin;PDBTitle: streptococcus pneumonia tigr4 flavodoxin: structural and biophysical2 characterization of a novel drug target
72 c3f6sI_
not modelled
98.6
15
PDB header: electron transportChain: I: PDB Molecule: flavodoxin;PDBTitle: desulfovibrio desulfuricans (atcc 29577) oxidized flavodoxin alternate2 conformers
73 c4heqB_
not modelled
98.5
15
PDB header: electron transportChain: B: PDB Molecule: flavodoxin;PDBTitle: the crystal structure of flavodoxin from desulfovibrio gigas
74 c5b3kA_
not modelled
98.4
13
PDB header: electron transportChain: A: PDB Molecule: uncharacterized protein pa3435;PDBTitle: c101a mutant of flavodoxin from pseudomonas aeruginosa
75 d1czna_
not modelled
98.3
15
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
76 d1oboa_
not modelled
98.3
15
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
77 c2mtbA_
not modelled
98.2
17
PDB header: electron transportChain: A: PDB Molecule: flavodoxin-2;PDBTitle: solution structure of apo_fldb
78 d1ag9a_
not modelled
98.2
14
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
79 c4oxxA_
not modelled
98.2
18
PDB header: electron transportChain: A: PDB Molecule: cindoxin;PDBTitle: crystal structure of cindoxin, surface entropy reduction mutant
80 d1tlla2
not modelled
98.2
13
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Cytochrome p450 reductase N-terminal domain-like
81 d1yoba1
not modelled
98.1
11
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
82 d2fcra_
not modelled
98.0
12
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
83 d1f4pa_
not modelled
98.0
18
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
84 c6ohkA_
not modelled
97.9
16
PDB header: electron transportChain: A: PDB Molecule: flavodoxin;PDBTitle: crystal structure of fusobacterium nucleatum flavodoxin mutant k13g2 bound to flavin mononucleotide
85 c2hnbA_
not modelled
97.9
19
PDB header: electron transportChain: A: PDB Molecule: protein mioc;PDBTitle: solution structure of a bacterial holo-flavodoxin
86 c4h2dB_
not modelled
97.9
13
PDB header: oxidoreductaseChain: B: PDB Molecule: nadph-dependent diflavin oxidoreductase 1;PDBTitle: crystal structure of ndor1
87 d1ykga1
not modelled
97.9
13
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Cytochrome p450 reductase N-terminal domain-like
88 c1bvyF_
not modelled
97.9
15
PDB header: oxidoreductaseChain: F: PDB Molecule: protein (cytochrome p450 bm-3);PDBTitle: complex of the heme and fmn-binding domains of the2 cytochrome p450(bm-3)
89 d1bvyf_
not modelled
97.9
15
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
90 c2wc1A_
not modelled
97.8
11
PDB header: electron transportChain: A: PDB Molecule: flavodoxin;PDBTitle: three-dimensional structure of the nitrogen fixation2 flavodoxin (niff) from rhodobacter capsulatus at 2.2 a
91 c2m6rA_
not modelled
97.8
12
PDB header: electron transportChain: A: PDB Molecule: flavodoxin;PDBTitle: apo_yqca
92 d1b1ca_
not modelled
97.8
18
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Cytochrome p450 reductase N-terminal domain-like
93 d1ja1a2
not modelled
97.7
20
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Cytochrome p450 reductase N-terminal domain-like
94 c3hr4C_
not modelled
97.5
15
PDB header: oxidoreductase/metal binding proteinChain: C: PDB Molecule: nitric oxide synthase, inducible;PDBTitle: human inos reductase and calmodulin complex
95 d1fuea_
not modelled
97.4
15
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related
96 c4n82B_
not modelled
97.2
14
PDB header: oxidoreductaseChain: B: PDB Molecule: ribonucleotide reductase;PDBTitle: x-ray crystal structure of streptococcus sanguinis nrdiox
97 c6efvA_
not modelled
96.1
15
PDB header: flavoproteinChain: A: PDB Molecule: sulfite reductase [nadph] flavoprotein alpha-component;PDBTitle: the nadph-dependent sulfite reductase flavoprotein adopts an extended2 conformation that is unique to this diflavin reductase
98 c5gxuA_
not modelled
94.9
13
PDB header: oxidoreductaseChain: A: PDB Molecule: nadph--cytochrome p450 reductase 2;PDBTitle: cystal structure of arabidopsis atr2
99 c1tllA_
not modelled
94.0
15
PDB header: oxidoreductaseChain: A: PDB Molecule: nitric-oxide synthase, brain;PDBTitle: crystal structure of rat neuronal nitric-oxide synthase2 reductase module at 2.3 a resolution.
100 d1n57a_
not modelled
93.5
13
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: DJ-1/PfpI
101 c2x6rA_
not modelled
92.8
11
PDB header: isomeraseChain: A: PDB Molecule: trehalose-synthase tret;PDBTitle: crystal structure of trehalose synthase tret from p.2 horikoshi produced by soaking in trehalose
102 c3c4vB_
not modelled
92.0
17
PDB header: transferaseChain: B: PDB Molecule: predicted glycosyltransferases;PDBTitle: structure of the retaining glycosyltransferase msha:the2 first step in mycothiol biosynthesis. organism:3 corynebacterium glutamicum : complex with udp and 1l-ins-1-4 p.
103 c1j9zB_
not modelled
91.4
19
PDB header: oxidoreductaseChain: B: PDB Molecule: nadph-cytochrome p450 reductase;PDBTitle: cypor-w677g
104 c2xmpB_
not modelled
90.7
11
PDB header: sugar binding proteinChain: B: PDB Molecule: trehalose-synthase tret;PDBTitle: crystal structure of trehalose synthase tret mutant e326a2 from p.horishiki in complex with udp
105 d1u7za_
not modelled
89.1
15
Fold: Ribokinase-likeSuperfamily: CoaB-likeFamily: CoaB-like
106 c4qjiB_
not modelled
88.6
19
PDB header: ligaseChain: B: PDB Molecule: phosphopantothenate--cysteine ligase;PDBTitle: crystal structure of the c-terminal ctp-binding domain of a2 phosphopantothenoylcysteine decarboxylase/phosphopantothenate-3 cysteine ligase with bound ctp from mycobacterium smegmatis
107 c2ofpB_
not modelled
87.9
8
PDB header: oxidoreductaseChain: B: PDB Molecule: ketopantoate reductase;PDBTitle: crystal structure of escherichia coli ketopantoate2 reductase in a ternary complex with nadp+ and pantoate
108 d2bisa1
not modelled
87.2
11
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: Glycosyl transferases group 1
109 c2bpoA_
not modelled
86.9
13
PDB header: reductaseChain: A: PDB Molecule: nadph-cytochrom p450 reductase;PDBTitle: crystal structure of the yeast cpr triple mutant: d74g, y75f, k78a.
110 d1ks9a2
not modelled
86.5
8
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain
111 c5n80A_
not modelled
86.2
7
PDB header: transferaseChain: A: PDB Molecule: lipopolysaccharide 1,6-galactosyltransferase;PDBTitle: glycosyltransferase lps biosynthesis in complex with udp
112 c3vpbC_
not modelled
85.9
11
PDB header: ligaseChain: C: PDB Molecule: putative acetylornithine deacetylase;PDBTitle: argx from sulfolobus tokodaii complexed with2 lysw/glu/adp/mg/zn/sulfate
113 c3d8tB_
not modelled
84.7
12
PDB header: lyaseChain: B: PDB Molecule: uroporphyrinogen-iii synthase;PDBTitle: thermus thermophilus uroporphyrinogen iii synthase
114 c3c7cB_
not modelled
84.2
11
PDB header: oxidoreductaseChain: B: PDB Molecule: octopine dehydrogenase;PDBTitle: a structural basis for substrate and stereo selectivity in2 octopine dehydrogenase (odh-nadh-l-arginine)
115 c6d9tA_
not modelled
84.1
8
PDB header: transferaseChain: A: PDB Molecule: glycosyl transferase;PDBTitle: bsha from staphylococcus aureus complexed with udp
116 c3ic5A_
not modelled
84.0
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative saccharopine dehydrogenase;PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
117 c2x2oA_
not modelled
83.8
20
PDB header: flavoproteinChain: A: PDB Molecule: nrdi protein;PDBTitle: the flavoprotein nrdi from bacillus cereus with the initially oxidized2 fmn cofactor in an intermediate radiation reduced state
118 d1mkza_
not modelled
83.4
22
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like
119 c3vufA_
not modelled
83.1
10
PDB header: transferaseChain: A: PDB Molecule: granule-bound starch synthase 1,PDBTitle: crystal structure of rice granule bound starch synthase i catalytic2 domain in complex with adp
120 c3lwzC_
not modelled
82.9
17
PDB header: lyaseChain: C: PDB Molecule: 3-dehydroquinate dehydratase;PDBTitle: 1.65 angstrom resolution crystal structure of type ii 3-dehydroquinate2 dehydratase (aroq) from yersinia pestis