| 1 |
|
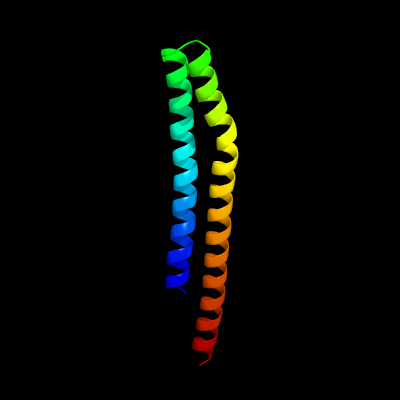
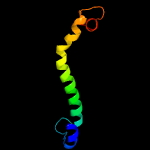
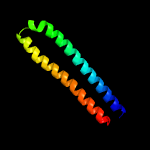
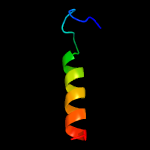
PDB 3zbh chain C
Region: 34 - 120
Aligned: 87
Modelled: 87
Confidence: 96.2%
Identity: 15%
PDB header:unknown function
Chain: C: PDB Molecule:esxa;
PDBTitle: geobacillus thermodenitrificans esxa crystal form i
Phyre2
| 2 |
|
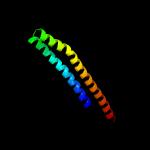
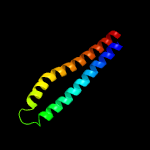
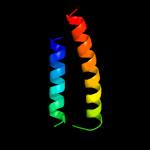
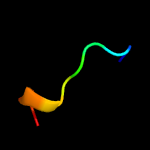
PDB 1wa8 chain A domain 1
Region: 34 - 122
Aligned: 89
Modelled: 89
Confidence: 95.8%
Identity: 18%
Fold: Ferritin-like
Superfamily: EsxAB dimer-like
Family: ESAT-6 like
Phyre2
| 3 |
|
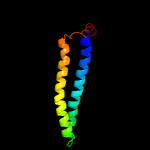
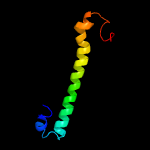
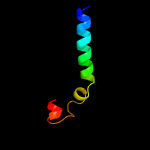
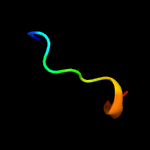
PDB 3gvm chain A
Region: 34 - 120
Aligned: 87
Modelled: 87
Confidence: 95.5%
Identity: 13%
PDB header:viral protein
Chain: A: PDB Molecule:putative uncharacterized protein sag1039;
PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
Phyre2
| 4 |
|
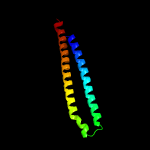
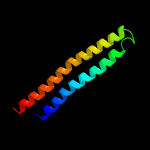
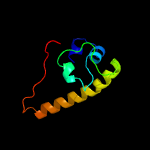
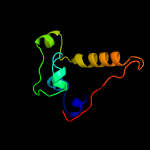
PDB 4lws chain A
Region: 30 - 123
Aligned: 94
Modelled: 94
Confidence: 95.3%
Identity: 11%
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
Phyre2
| 5 |
|
PDB 2kg7 chain B
Region: 61 - 121
Aligned: 61
Modelled: 61
Confidence: 88.9%
Identity: 11%
PDB header:unknown function
Chain: B: PDB Molecule:esat-6-like protein esxh;
PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
Phyre2
| 6 |
|
PDB 2vs0 chain B
Region: 34 - 111
Aligned: 78
Modelled: 78
Confidence: 87.9%
Identity: 10%
PDB header:cell invasion
Chain: B: PDB Molecule:virulence factor esxa;
PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
Phyre2
| 7 |
|
PDB 1wa8 chain B domain 1
Region: 61 - 121
Aligned: 61
Modelled: 61
Confidence: 86.4%
Identity: 18%
Fold: Ferritin-like
Superfamily: EsxAB dimer-like
Family: ESAT-6 like
Phyre2
| 8 |
|
PDB 4iog chain D
Region: 34 - 111
Aligned: 78
Modelled: 78
Confidence: 80.1%
Identity: 14%
PDB header:unknown function
Chain: D: PDB Molecule:secreted protein esxb;
PDBTitle: the crystal structure of a secreted protein esxb (wild-type, in p212 space group) from bacillus anthracis str. sterne
Phyre2
| 9 |
|
PDB 4lws chain B
Region: 30 - 109
Aligned: 80
Modelled: 80
Confidence: 63.1%
Identity: 18%
PDB header:unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
Phyre2
| 10 |
|
PDB 4i0x chain J
Region: 49 - 97
Aligned: 49
Modelled: 49
Confidence: 42.1%
Identity: 18%
PDB header:structural genomics, unknown function
Chain: J: PDB Molecule:esat-6-like protein mab_3113;
PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
Phyre2
| 11 |
|
PDB 2fef chain A domain 1
Region: 77 - 113
Aligned: 37
Modelled: 37
Confidence: 13.5%
Identity: 30%
Fold: Bromodomain-like
Superfamily: PA2201 N-terminal domain-like
Family: PA2201 N-terminal domain-like
Phyre2
| 12 |
|
PDB 4xwh chain A
Region: 35 - 112
Aligned: 70
Modelled: 78
Confidence: 11.4%
Identity: 26%
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-n-acetylglucosaminidase;
PDBTitle: crystal structure of the human n-acetyl-alpha-glucosaminidase
Phyre2
| 13 |
|
PDB 2p7v chain A
Region: 78 - 92
Aligned: 15
Modelled: 15
Confidence: 11.3%
Identity: 20%
PDB header:transcription
Chain: A: PDB Molecule:regulator of sigma d;
PDBTitle: crystal structure of the escherichia coli regulator of sigma 70, rsd,2 in complex with sigma 70 domain 4
Phyre2
| 14 |
|
PDB 6icz chain X
Region: 70 - 95
Aligned: 26
Modelled: 26
Confidence: 8.8%
Identity: 35%
PDB header:splicing
Chain: X: PDB Molecule:prkr-interacting protein 1;
PDBTitle: cryo-em structure of a human post-catalytic spliceosome (p complex) at2 3.0 angstrom
Phyre2
| 15 |
|
PDB 5vko chain B
Region: 11 - 17
Aligned: 7
Modelled: 7
Confidence: 8.8%
Identity: 86%
PDB header:transcription
Chain: B: PDB Molecule:dna-directed rna polymerase ii subunit rpb1;
PDBTitle: spt6 tsh2-rpb1 1468-1500 pt1471, ps1493
Phyre2
| 16 |
|
PDB 1v8d chain A
Region: 95 - 104
Aligned: 10
Modelled: 10
Confidence: 7.8%
Identity: 60%
Fold: TTHA0583/YokD-like
Superfamily: TTHA0583/YokD-like
Family: Hypothetical protein TT1679
Phyre2
| 17 |
|
PDB 1v8d chain C
Region: 95 - 104
Aligned: 10
Modelled: 10
Confidence: 7.6%
Identity: 60%
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:hypothetical protein (tt1679);
PDBTitle: crystal structure of the conserved hypothetical protein2 tt1679 from thermus thermophilus
Phyre2
| 18 |
|
PDB 2vcb chain A
Region: 35 - 112
Aligned: 70
Modelled: 78
Confidence: 6.9%
Identity: 20%
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-n-acetylglucosaminidase;
PDBTitle: family 89 glycoside hydrolase from clostridium perfringens2 in complex with pugnac
Phyre2
| 19 |
|
PDB 3faj chain A
Region: 33 - 45
Aligned: 13
Modelled: 13
Confidence: 6.6%
Identity: 54%
PDB header:structural protein
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: structure of the structural protein p131 of the archaeal virus2 acidianus two-tailed virus (atv)
Phyre2
| 20 |
|
PDB 1v7r chain A
Region: 107 - 119
Aligned: 13
Modelled: 13
Confidence: 6.2%
Identity: 62%
Fold: Anticodon-binding domain-like
Superfamily: ITPase-like
Family: ITPase (Ham1)
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|