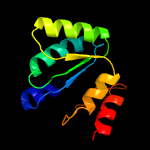
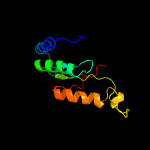
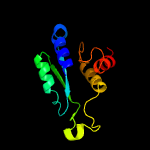
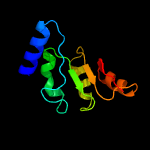
1 c3gndC_
94.4
14
PDB header: lyaseChain: C: PDB Molecule: aldolase lsrf;PDBTitle: crystal structure of e. coli lsrf in complex with ribulose-5-phosphate
2 d1ojxa_
93.3
15
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
3 c2qjhH_
87.7
17
PDB header: lyaseChain: H: PDB Molecule: putative aldolase mj0400;PDBTitle: m. jannaschii adh synthase covalently bound to2 dihydroxyacetone phosphate
4 c1rr2A_
86.0
15
PDB header: transferaseChain: A: PDB Molecule: transcarboxylase 5s subunit;PDBTitle: propionibacterium shermanii transcarboxylase 5s subunit bound to 2-2 ketobutyric acid
5 c5n2pA_
85.5
15
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: sulfolobus solfataricus tryptophan synthase a
6 c5kzmA_
84.2
25
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: crystal structure of tryptophan synthase alpha-beta chain complex from2 francisella tularensis
7 c4lrtC_
83.3
27
PDB header: lyase/oxidoreductaseChain: C: PDB Molecule: 4-hydroxy-2-oxovalerate aldolase;PDBTitle: crystal and solution structures of the bifunctional enzyme2 (aldolase/aldehyde dehydrogenase) from thermomonospora curvata,3 reveal a cofactor-binding domain motion during nad+ and coa4 accommodation whithin the shared cofactor-binding site
8 c3tfxB_
81.8
17
PDB header: lyaseChain: B: PDB Molecule: orotidine 5'-phosphate decarboxylase;PDBTitle: crystal structure of orotidine 5'-phosphate decarboxylase from2 lactobacillus acidophilus
9 c4euhA_
81.5
26
PDB header: oxidoreductaseChain: A: PDB Molecule: putative reductase ca_c0462;PDBTitle: crystal structure of clostridium acetobutulicum trans-2-enoyl-coa2 reductase apo form
10 c5ks8F_
79.2
20
PDB header: ligaseChain: F: PDB Molecule: pyruvate carboxylase subunit beta;PDBTitle: crystal structure of two-subunit pyruvate carboxylase from2 methylobacillus flagellatus
11 c3bg3A_
76.5
26
PDB header: ligaseChain: A: PDB Molecule: pyruvate carboxylase, mitochondrial;PDBTitle: crystal structure of human pyruvate carboxylase (missing the biotin2 carboxylase domain at the n-terminus)
12 d1o94c_
74.6
15
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits
13 c4jn6C_
74.1
24
PDB header: lyase/oxidoreductaseChain: C: PDB Molecule: 4-hydroxy-2-oxovalerate aldolase;PDBTitle: crystal structure of the aldolase-dehydrogenase complex from2 mycobacterium tuberculosis hrv37
14 c5ey5A_
73.8
19
PDB header: lyaseChain: A: PDB Molecule: lbcats-a;PDBTitle: lbcats
15 c1ydoC_
70.7
23
PDB header: lyaseChain: C: PDB Molecule: hmg-coa lyase;PDBTitle: crystal structure of the bacillis subtilis hmg-coa lyase, northeast2 structural genomics target sr181.
16 d1t1ra2
70.1
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
17 c3bg3B_
70.0
14
PDB header: ligaseChain: B: PDB Molecule: pyruvate carboxylase, mitochondrial;PDBTitle: crystal structure of human pyruvate carboxylase (missing the biotin2 carboxylase domain at the n-terminus)
18 d1efpb_
69.9
18
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits
19 c4nu7C_
69.4
18
PDB header: isomeraseChain: C: PDB Molecule: ribulose-phosphate 3-epimerase;PDBTitle: 2.05 angstrom crystal structure of ribulose-phosphate 3-epimerase from2 toxoplasma gondii.
20 c3h75A_
68.3
20
PDB header: sugar binding proteinChain: A: PDB Molecule: periplasmic sugar-binding domain protein;PDBTitle: crystal structure of a periplasmic sugar-binding protein from the2 pseudomonas fluorescens
21 c1k5hB_
not modelled
67.5
15
PDB header: oxidoreductaseChain: B: PDB Molecule: 1-deoxy-d-xylulose-5-phosphate reductoisomerase;PDBTitle: 1-deoxy-d-xylulose-5-phosphate reductoisomerase
22 d2fywa1
not modelled
67.5
23
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like
23 c3ct7E_
not modelled
66.7
16
PDB header: isomeraseChain: E: PDB Molecule: d-allulose-6-phosphate 3-epimerase;PDBTitle: crystal structure of d-allulose 6-phosphate 3-epimerase2 from escherichia coli k-12
24 c4go4E_
not modelled
66.4
21
PDB header: oxidoreductaseChain: E: PDB Molecule: putative gamma-hydroxymuconic semialdehyde dehydrogenase;PDBTitle: crystal structure of pnpe in complex with nicotinamide adenine2 dinucleotide
25 d1tqxa_
not modelled
66.3
12
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: D-ribulose-5-phosphate 3-epimerase
26 c2ekcA_
not modelled
66.2
18
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
27 d1qopa_
not modelled
65.7
20
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes
28 c4kpuB_
not modelled
65.3
17
PDB header: electron transportChain: B: PDB Molecule: electron transfer flavoprotein alpha/beta-subunit;PDBTitle: electron transferring flavoprotein of acidaminococcus fermentans:2 towards a mechanism of flavin-based electron bifurcation
29 c3zu3A_
not modelled
65.0
28
PDB header: oxidoreductaseChain: A: PDB Molecule: putative reductase ypo4104/y4119/yp_4011;PDBTitle: structure of the enoyl-acp reductase fabv from yersinia pestis with2 the cofactor nadh (mr, cleaved histag)
30 c3ed6B_
not modelled
64.1
22
PDB header: oxidoreductaseChain: B: PDB Molecule: betaine aldehyde dehydrogenase;PDBTitle: 1.7 angstrom resolution crystal structure of betaine aldehyde2 dehydrogenase (betb) from staphylococcus aureus
31 c5ks8D_
not modelled
64.1
19
PDB header: ligaseChain: D: PDB Molecule: pyruvate carboxylase subunit beta;PDBTitle: crystal structure of two-subunit pyruvate carboxylase from2 methylobacillus flagellatus
32 c3a9iA_
not modelled
63.9
21
PDB header: transferase/transferase inhibitorChain: A: PDB Molecule: homocitrate synthase;PDBTitle: crystal structure of homocitrate synthase from thermus thermophilus2 complexed with lys
33 c3bfjK_
not modelled
63.8
25
PDB header: oxidoreductaseChain: K: PDB Molecule: 1,3-propanediol oxidoreductase;PDBTitle: crystal structure analysis of 1,3-propanediol oxidoreductase
34 c4k05B_
not modelled
63.0
24
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: conserved hypothetical exported protein;PDBTitle: crystal structure of a duf1343 family protein (bf0371) from2 bacteroides fragilis nctc 9343 at 1.65 a resolution
35 c4qslC_
not modelled
62.2
20
PDB header: ligaseChain: C: PDB Molecule: pyruvate carboxylase;PDBTitle: crystal structure of listeria monocytogenes pyruvate carboxylase
36 d1h1ya_
not modelled
61.9
12
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: D-ribulose-5-phosphate 3-epimerase
37 c5xi0B_
not modelled
61.6
31
PDB header: oxidoreductaseChain: B: PDB Molecule: enoyl-[acyl-carrier-protein] reductase [nadh];PDBTitle: crystal structure of fabv, a new class of enyl-acyl carrier protein2 reductase from vibrio fischeri
38 d1rqba2
not modelled
60.8
19
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: HMGL-like
39 c3navB_
not modelled
59.8
25
PDB header: lyaseChain: B: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
40 c3i44A_
not modelled
59.8
19
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase;PDBTitle: crystal structure of aldehyde dehydrogenase from bartonella2 henselae at 2.0a resolution
41 c5kqoA_
not modelled
58.2
17
PDB header: oxidoreductaseChain: A: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: 1-deoxy-d-xylulose 5-phosphate reductoisomerase from vibrio vulnificus
42 d1pvda1
not modelled
57.3
13
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
43 d1r0ka2
not modelled
56.5
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
44 c3lvuB_
not modelled
55.9
20
PDB header: transport proteinChain: B: PDB Molecule: abc transporter, periplasmic substrate-binding protein;PDBTitle: crystal structure of abc transporter, periplasmic substrate-binding2 protein spo2066 from silicibacter pomeroyi
45 d1xcfa_
not modelled
55.8
13
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes
46 c5ol2E_
not modelled
55.6
17
PDB header: flavoproteinChain: E: PDB Molecule: electron transfer flavoprotein small subunit;PDBTitle: the electron transferring flavoprotein/butyryl-coa dehydrogenase2 complex from clostridium difficile
47 c4ov9A_
not modelled
55.0
19
PDB header: transferaseChain: A: PDB Molecule: isopropylmalate synthase;PDBTitle: structure of isopropylmalate synthase binding with alpha-2 isopropylmalate
48 c3thaB_
not modelled
54.9
15
PDB header: lyaseChain: B: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: tryptophan synthase subunit alpha from campylobacter jejuni.
49 c3rmjB_
not modelled
54.8
19
PDB header: transferaseChain: B: PDB Molecule: 2-isopropylmalate synthase;PDBTitle: crystal structure of truncated alpha-isopropylmalate synthase from2 neisseria meningitidis
50 d1dvja_
not modelled
54.3
22
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Decarboxylase
51 c3zs6A_
not modelled
54.2
16
PDB header: peptide binding proteinChain: A: PDB Molecule: periplasmic oligopeptide-binding protein;PDBTitle: the structural characterization of burkholderia pseudomallei oppa.
52 d1rd5a_
not modelled
53.4
13
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes
53 c3ru6C_
not modelled
53.2
13
PDB header: lyaseChain: C: PDB Molecule: orotidine 5'-phosphate decarboxylase;PDBTitle: 1.8 angstrom resolution crystal structure of orotidine 5'-phosphate2 decarboxylase (pyrf) from campylobacter jejuni subsp. jejuni nctc3 11168
54 c4qslE_
not modelled
52.9
19
PDB header: ligaseChain: E: PDB Molecule: pyruvate carboxylase;PDBTitle: crystal structure of listeria monocytogenes pyruvate carboxylase
55 d1nvma2
not modelled
52.7
16
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: HMGL-like
56 c3l55B_
not modelled
52.7
9
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: b-1,4-endoglucanase/cellulase;PDBTitle: crystal structure of a putative beta-1,4-endoglucanase / cellulase2 from prevotella bryantii
57 d2amxa1
not modelled
51.7
18
Fold: TIM beta/alpha-barrelSuperfamily: Metallo-dependent hydrolasesFamily: Adenosine/AMP deaminase
58 c3rmsA_
not modelled
50.7
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein svir_20580 from2 saccharomonospora viridis
59 d3clsc1
not modelled
50.4
14
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits
60 c2d4eB_
not modelled
50.3
25
PDB header: oxidoreductaseChain: B: PDB Molecule: 5-carboxymethyl-2-hydroxymuconate semialdehydePDBTitle: crystal structure of the hpcc from thermus thermophilus hb8
61 d1x7fa2
not modelled
50.0
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Outer surface protein, N-terminal domain
62 c3p96A_
not modelled
49.9
24
PDB header: hydrolaseChain: A: PDB Molecule: phosphoserine phosphatase serb;PDBTitle: crystal structure of phosphoserine phosphatase serb from mycobacterium2 avium, native form
63 d1gyta2
not modelled
49.7
21
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: Leucine aminopeptidase, C-terminal domain
64 d1vlja_
not modelled
49.4
17
Fold: Dehydroquinate synthase-likeSuperfamily: Dehydroquinate synthase-likeFamily: Iron-containing alcohol dehydrogenase
65 c1r0lD_
not modelled
49.2
19
PDB header: oxidoreductaseChain: D: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: 1-deoxy-d-xylulose 5-phosphate reductoisomerase from zymomonas mobilis2 in complex with nadph
66 c3ivuB_
not modelled
48.3
20
PDB header: transferaseChain: B: PDB Molecule: homocitrate synthase, mitochondrial;PDBTitle: homocitrate synthase lys4 bound to 2-og
67 c1ta9A_
not modelled
47.8
13
PDB header: oxidoreductaseChain: A: PDB Molecule: glycerol dehydrogenase;PDBTitle: crystal structure of glycerol dehydrogenase from schizosaccharomyces2 pombe
68 c1mwoA_
not modelled
47.5
30
PDB header: hydrolaseChain: A: PDB Molecule: alpha amylase;PDBTitle: crystal structure analysis of the hyperthermostable2 pyrocoocus woesei alpha-amylase
69 c2cw6B_
not modelled
46.2
16
PDB header: lyaseChain: B: PDB Molecule: hydroxymethylglutaryl-coa lyase, mitochondrial;PDBTitle: crystal structure of human hmg-coa lyase: insights into2 catalysis and the molecular basis for3 hydroxymethylglutaric aciduria
70 c1ydnA_
not modelled
46.2
25
PDB header: lyaseChain: A: PDB Molecule: hydroxymethylglutaryl-coa lyase;PDBTitle: crystal structure of the hmg-coa lyase from brucella melitensis,2 northeast structural genomics target lr35.
71 c2nx9B_
not modelled
46.0
19
PDB header: lyaseChain: B: PDB Molecule: oxaloacetate decarboxylase 2, subunit alpha;PDBTitle: crystal structure of the carboxyltransferase domain of the2 oxaloacetate decarboxylase na+ pump from vibrio cholerae
72 d1uuqa_
not modelled
45.8
29
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
73 c1uz4A_
not modelled
45.8
29
PDB header: hydrolaseChain: A: PDB Molecule: man5a;PDBTitle: common inhibition of beta-glucosidase and beta-mannosidase2 by isofagomine lactam reflects different conformational3 intineraries for glucoside and mannoside hydrolysis
74 c4hnvB_
not modelled
45.8
20
PDB header: ligaseChain: B: PDB Molecule: pyruvate carboxylase;PDBTitle: crystal structure of r54e mutant of s. aureus pyruvate carboxylase
75 d1efvb_
not modelled
45.8
19
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits
76 d1dbta_
not modelled
45.5
15
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Decarboxylase
77 c3qr3B_
not modelled
45.4
17
PDB header: hydrolaseChain: B: PDB Molecule: endoglucanase eg-ii;PDBTitle: crystal structure of cel5a (eg2) from hypocrea jecorina (trichoderma2 reesei)
78 c3icgD_
not modelled
45.2
19
PDB header: hydrolaseChain: D: PDB Molecule: endoglucanase d;PDBTitle: crystal structure of the catalytic and carbohydrate binding domain of2 endoglucanase d from clostridium cellulovorans
79 c2oylB_
not modelled
45.1
26
PDB header: hydrolaseChain: B: PDB Molecule: endoglycoceramidase ii;PDBTitle: endo-glycoceramidase ii from rhodococcus sp.: cellobiose-like2 imidazole complex
80 c3ff4A_
not modelled
45.0
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein chu_1412
81 c6gl2A_
not modelled
44.8
15
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase, family gh5;PDBTitle: structure of zgengagh5_4 wild type at 1.2 angstrom resolution
82 c3ox4D_
not modelled
44.5
23
PDB header: oxidoreductaseChain: D: PDB Molecule: alcohol dehydrogenase 2;PDBTitle: structures of iron-dependent alcohol dehydrogenase 2 from zymomonas2 mobilis zm4 complexed with nad cofactor
83 c3iwkB_
not modelled
44.1
18
PDB header: oxidoreductaseChain: B: PDB Molecule: aminoaldehyde dehydrogenase;PDBTitle: crystal structure of aminoaldehyde dehydrogenase 1 from pisum sativum2 (psamadh1)
84 c5vbfH_
not modelled
43.9
17
PDB header: oxidoreductaseChain: H: PDB Molecule: nad-dependent succinate-semialdehyde dehydrogenase;PDBTitle: crystal structure of succinate semialdehyde dehydrogenase from2 burkholderia vietnamiensis
85 d1q6za1
not modelled
43.9
18
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
86 c4zn6B_
not modelled
43.7
14
PDB header: oxidoreductaseChain: B: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: x-ray crystal structure of 1-deoxy-d-xylulose 5-phosphate2 reductoisomerase (ispc) from acinetobacter baumannii
87 c4dbeB_
not modelled
43.6
14
PDB header: lyase/lyase inhibitorChain: B: PDB Molecule: orotidine 5'-phosphate decarboxylase;PDBTitle: crystal structure of orotidine 5'-monophosphate decarboxylase from2 sulfolobus solfataricus complexed with inhibitor bmp
88 c4df1B_
not modelled
43.4
38
PDB header: lyase/lyase inhibitorChain: B: PDB Molecule: orotidine 5'-phosphate decarboxylase;PDBTitle: crystal structure of orotidine 5'-monophosphate decarboxylase from2 thermoproteus neutrophilus complexed with inhibitor bmp
89 d1rrma_
not modelled
43.4
18
Fold: Dehydroquinate synthase-likeSuperfamily: Dehydroquinate synthase-likeFamily: Iron-containing alcohol dehydrogenase
90 d1wa3a1
not modelled
43.3
19
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
91 c1nvmG_
not modelled
43.2
16
PDB header: lyase/oxidoreductaseChain: G: PDB Molecule: 4-hydroxy-2-oxovalerate aldolase;PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
92 c5e09A_
not modelled
42.8
19
PDB header: hydrolaseChain: A: PDB Molecule: cellulase;PDBTitle: structural insight of a trimodular halophilic cellulase with a family2 46 carbohydrate-binding module
93 c3ncoA_
not modelled
42.8
16
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase fncel5a;PDBTitle: crystal structure of fncel5a from f. nodosum rt17-b1
94 d2djia1
not modelled
41.9
18
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
95 c3vndD_
not modelled
41.9
20
PDB header: lyaseChain: D: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: crystal structure of tryptophan synthase alpha-subunit from the2 psychrophile shewanella frigidimarina k14-2
96 c2q5cA_
not modelled
41.6
18
PDB header: transcriptionChain: A: PDB Molecule: ntrc family transcriptional regulator;PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
97 d1rxwa2
not modelled
41.5
7
Fold: PIN domain-likeSuperfamily: PIN domain-likeFamily: 5' to 3' exonuclease catalytic domain
98 c5ow0B_
not modelled
41.4
22
PDB header: electron transportChain: B: PDB Molecule: electron transfer flavoprotein, beta subunit;PDBTitle: crystal structure of an electron transfer flavoprotein from2 geobacillus metallireducens
99 d1qhoa4
not modelled
41.3
26
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
100 c3a24A_
not modelled
41.2
24
PDB header: hydrolaseChain: A: PDB Molecule: alpha-galactosidase;PDBTitle: crystal structure of bt1871 retaining glycosidase
101 c2yytA_
not modelled
40.8
31
PDB header: lyaseChain: A: PDB Molecule: orotidine 5'-phosphate decarboxylase;PDBTitle: crystal structure of uncharacterized conserved protein from2 geobacillus kaustophilus
102 c3dmyA_
not modelled
40.7
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein fdra;PDBTitle: crystal structure of a predicated acyl-coa synthetase from e.coli
103 c2vp8A_
not modelled
40.5
18
PDB header: transferaseChain: A: PDB Molecule: dihydropteroate synthase 2;PDBTitle: structure of mycobacterium tuberculosis rv1207
104 d1losc_
not modelled
40.4
20
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Decarboxylase
105 c4yztA_
not modelled
40.4
19
PDB header: hydrolaseChain: A: PDB Molecule: cellulose hydrolase;PDBTitle: crystal structure of a tri-modular gh5 (subfamily 4) endo-beta-1, 4-2 glucanase from bacillus licheniformis complexed with cellotetraose
106 c3igfB_
not modelled
40.4
17
PDB header: atp binding proteinChain: B: PDB Molecule: all4481 protein;PDBTitle: crystal structure of the all4481 protein from nostoc sp. pcc 7120,2 northeast structural genomics consortium target nsr300
107 c5ccuA_
not modelled
40.1
15
PDB header: hydrolaseChain: A: PDB Molecule: putative secreted endoglycosylceramidase;PDBTitle: crystal structure of endoglycoceramidase i from rhodococ-cus equi
108 c3ldvB_
not modelled
40.0
31
PDB header: lyaseChain: B: PDB Molecule: orotidine 5'-phosphate decarboxylase;PDBTitle: 1.77 angstrom resolution crystal structure of orotidine 5'-phosphate2 decarboxylase from vibrio cholerae o1 biovar eltor str. n16961
109 c3urkA_
not modelled
39.6
16
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase;PDBTitle: isph in complex with propynyl diphosphate (1061)
110 c3ke8A_
not modelled
39.6
16
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase;PDBTitle: crystal structure of isph:hmbpp-complex
111 c2eghA_
not modelled
39.6
14
PDB header: oxidoreductaseChain: A: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase2 complexed with a magnesium ion, nadph and fosmidomycin
112 c3pamB_
not modelled
39.4
24
PDB header: transport proteinChain: B: PDB Molecule: transmembrane protein;PDBTitle: crystal structure of a domain of transmembrane protein of abc-type2 oligopeptide transport system from bartonella henselae str. houston-1
113 d1ovma1
not modelled
39.4
15
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain
114 d1mkza_
not modelled
39.4
18
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like
115 c3jr2D_
not modelled
39.4
22
PDB header: biosynthetic proteinChain: D: PDB Molecule: hexulose-6-phosphate synthase sgbh;PDBTitle: x-ray crystal structure of the mg-bound 3-keto-l-gulonate-6-phosphate2 decarboxylase from vibrio cholerae o1 biovar el tor str. n16961
116 c3mmwB_
not modelled
39.0
18
PDB header: hydrolaseChain: B: PDB Molecule: endoglucanase;PDBTitle: crystal structure of endoglucanase cel5a from the hyperthermophilic2 thermotoga maritima
117 c4v2xA_
not modelled
38.9
19
PDB header: hydrolaseChain: A: PDB Molecule: endo-beta-1,4-glucanase (cellulase b);PDBTitle: high resolution structure of the full length tri-modular2 endo-beta-1,4-glucanase b (cel5b) from bacillus halodurans
118 c6hlxA_
not modelled
38.2
19
PDB header: transport proteinChain: A: PDB Molecule: agaa;PDBTitle: structure of the pbp agaa in complex with agropinic acid from2 a.tumefacien r10
119 c2fpgA_
not modelled
38.2
14
PDB header: ligaseChain: A: PDB Molecule: succinyl-coa ligase [gdp-forming] alpha-chain,PDBTitle: crystal structure of pig gtp-specific succinyl-coa synthetase in2 complex with gdp
120 c2zunB_
not modelled
38.0
19
PDB header: hydrolaseChain: B: PDB Molecule: 458aa long hypothetical endo-1,4-beta-glucanase;PDBTitle: functional analysis of hyperthermophilic endocellulase from2 the archaeon pyrococcus horikoshii