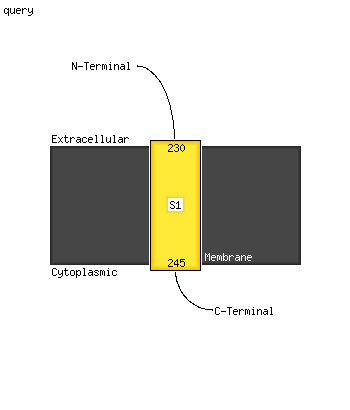
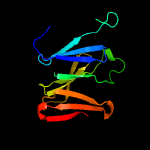
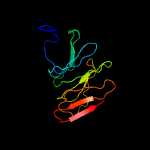
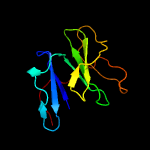
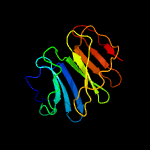
1 c1qniE_
99.8
21
PDB header: oxidoreductaseChain: E: PDB Molecule: nitrous-oxide reductase;PDBTitle: crystal structure of nitrous oxide reductase from pseudomonas nautica,2 at 2.4a resolution
2 c1fwxB_
99.8
11
PDB header: oxidoreductaseChain: B: PDB Molecule: nitrous oxide reductase;PDBTitle: crystal structure of nitrous oxide reductase from p. denitrificans
3 c3s8vA_
99.8
14
PDB header: signaling proteinChain: A: PDB Molecule: low-density lipoprotein receptor-related protein 6;PDBTitle: crystal structure of lrp6-dkk1 complex
4 d1qnia2
99.7
17
Fold: 7-bladed beta-propellerSuperfamily: Nitrous oxide reductase, N-terminal domainFamily: Nitrous oxide reductase, N-terminal domain
5 c3bwsA_
99.7
26
PDB header: unknown functionChain: A: PDB Molecule: protein lp49;PDBTitle: crystal structure of the leptospiral antigen lp49
6 c3s94A_
99.7
13
PDB header: signaling proteinChain: A: PDB Molecule: low-density lipoprotein receptor-related protein 6;PDBTitle: crystal structure of lrp6-e1e2
7 c5i5iA_
99.7
24
PDB header: oxidoreductaseChain: A: PDB Molecule: nitrous-oxide reductase;PDBTitle: shewanella denitrificans nitrous oxide reductase, app form
8 c6gc1A_
99.7
22
PDB header: unknown functionChain: A: PDB Molecule: nhl repeat-containing protein 2;PDBTitle: crystal structure of trx-like and nhl repeat containing domains of2 human nhlrc2
9 c5f30B_
99.7
23
PDB header: oxidoreductaseChain: B: PDB Molecule: thiocyanate dehydrogenase;PDBTitle: thiocyanate dehydrogenase from thioalkalivibrio paradoxus
10 c3sbrF_
99.7
10
PDB header: oxidoreductaseChain: F: PDB Molecule: nitrous-oxide reductase;PDBTitle: pseudomonas stutzeri nitrous oxide reductase, p1 crystal form with2 substrate
11 c3u4yA_
99.7
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of a functionally unknown protein (dtox_1751)2 from desulfotomaculum acetoxidans dsm 771.
12 c6f0qB_
99.7
53
PDB header: de novo proteinChain: B: PDB Molecule: pizza6-ayw;PDBTitle: crystal structure of pizza6-ayw
13 d1fwxa2
99.6
12
Fold: 7-bladed beta-propellerSuperfamily: Nitrous oxide reductase, N-terminal domainFamily: Nitrous oxide reductase, N-terminal domain
14 c2iwkB_
99.6
20
PDB header: oxidoreductaseChain: B: PDB Molecule: nitrous oxide reductase;PDBTitle: inhibitor-bound form of nitrous oxide reductase from2 achromobacter cycloclastes at 1.7 angstrom resolution
15 c3hrpA_
99.6
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of structural genomics protein of unknown function2 (np_812590.1) from bacteroides thetaiotaomicron vpi-5482 at 1.70 a3 resolution
16 c4hw6D_
99.6
17
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: hypothetical protein, ipt/tig domain protein;PDBTitle: crystal structure of an auxiliary nutrient binding protein2 (bacova_00264) from bacteroides ovatus atcc 8483 at 1.70 a resolution
17 c1n7dA_
99.6
22
PDB header: lipid transportChain: A: PDB Molecule: low-density lipoprotein receptor;PDBTitle: extracellular domain of the ldl receptor
18 c1l0qC_
99.6
44
PDB header: protein bindingChain: C: PDB Molecule: surface layer protein;PDBTitle: tandem yvtn beta-propeller and pkd domains from an archaeal surface2 layer protein
19 c2iwaA_
99.6
12
PDB header: transferaseChain: A: PDB Molecule: glutamine cyclotransferase;PDBTitle: unbound glutaminyl cyclotransferase from carica papaya.
20 c1gq1B_
99.6
15
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome cd1 nitrite reductase;PDBTitle: cytochrome cd1 nitrite reductase, y25s mutant, oxidised form
21 d1jofa_
not modelled
99.6
20
Fold: 7-bladed beta-propellerSuperfamily: 3-carboxy-cis,cis-mucoante lactonizing enzymeFamily: 3-carboxy-cis,cis-mucoante lactonizing enzyme
22 c3kyaA_
not modelled
99.6
18
PDB header: hydrolaseChain: A: PDB Molecule: putative phosphatase;PDBTitle: crystal structure of putative phosphatase (np_812416.1) from2 bacteroides thetaiotaomicron vpi-5482 at 1.77 a resolution
23 c3vh0C_
not modelled
99.5
16
PDB header: protein binding/dnaChain: C: PDB Molecule: uncharacterized protein ynce;PDBTitle: crystal structure of e. coli ynce complexed with dna
24 c3nolA_
not modelled
99.5
11
PDB header: transferaseChain: A: PDB Molecule: glutamine cyclotransferase;PDBTitle: crystal structure of zymomonas mobilis glutaminyl cyclase (trigonal2 form)
25 c2z2pA_
not modelled
99.5
9
PDB header: lyase/antibioticChain: A: PDB Molecule: virginiamycin b lyase;PDBTitle: crystal structure of catalytically inactive h270a virginiamycin b2 lyase from staphylococcus aureus with quinupristin
26 c3mbrX_
not modelled
99.5
11
PDB header: transferaseChain: X: PDB Molecule: glutamine cyclotransferase;PDBTitle: crystal structure of the glutaminyl cyclase from xanthomonas2 campestris
27 d2p4oa1
not modelled
99.5
13
Fold: 6-bladed beta-propellerSuperfamily: Calcium-dependent phosphotriesteraseFamily: All0351-like
28 c6fayA_
not modelled
99.5
17
PDB header: cell adhesionChain: A: PDB Molecule: odz3 protein;PDBTitle: teneurin3 monomer
29 d2g38a1
not modelled
99.5
26
Fold: Ferritin-likeSuperfamily: PE/PPE dimer-likeFamily: PE
30 c2g38A_
not modelled
99.5
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: pe family protein;PDBTitle: a pe/ppe protein complex from mycobacterium tuberculosis
31 c6fb3A_
not modelled
99.5
14
PDB header: cell adhesionChain: A: PDB Molecule: teneurin-2;PDBTitle: teneurin 2 partial extracellular domain
32 c6rteB_
not modelled
99.5
16
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome c;PDBTitle: dihydro-heme d1 dehydrogenase nirn in complex with dhe
33 c5xfsA_
not modelled
99.5
43
PDB header: protein transportChain: A: PDB Molecule: pe family protein pe8;PDBTitle: crystal structure of pe8-ppe15 in complex with espg5 from m.2 tuberculosis
34 c1nnoA_
not modelled
99.5
13
PDB header: oxidoreductaseChain: A: PDB Molecule: nitrite reductase;PDBTitle: conformational changes occurring upon no binding in nitrite reductase2 from pseudomonas aeruginosa
35 d1jmxb_
not modelled
99.5
23
Fold: 7-bladed beta-propellerSuperfamily: YVTN repeat-like/Quinoprotein amine dehydrogenaseFamily: Quinohemoprotein amine dehydrogenase B chain
36 c3fw0A_
not modelled
99.5
27
PDB header: lyaseChain: A: PDB Molecule: peptidyl-glycine alpha-amidating monooxygenase;PDBTitle: structure of peptidyl-alpha-hydroxyglycine alpha-amidating2 lyase (pal) bound to alpha-hydroxyhippuric acid (non-3 peptidic substrate)
37 d1qksa2
not modelled
99.4
15
Fold: 8-bladed beta-propellerSuperfamily: C-terminal (heme d1) domain of cytochrome cd1-nitrite reductaseFamily: C-terminal (heme d1) domain of cytochrome cd1-nitrite reductase
38 d1pjxa_
not modelled
99.4
19
Fold: 6-bladed beta-propellerSuperfamily: Calcium-dependent phosphotriesteraseFamily: SGL-like
39 d1l0qa2
not modelled
99.4
44
Fold: 7-bladed beta-propellerSuperfamily: YVTN repeat-like/Quinoprotein amine dehydrogenaseFamily: YVTN repeat
40 c5c2vB_
not modelled
99.4
24
PDB header: oxidoreductaseChain: B: PDB Molecule: hydrazine synthase beta subunit;PDBTitle: kuenenia stuttgartiensis hydrazine synthase
41 c3hfqB_
not modelled
99.4
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein lp_2219;PDBTitle: crystal structure of the lp_2219 protein from lactobacillus plantarum.2 northeast structural genomics consortium target lpr118.
42 c5d9bA_
not modelled
99.4
18
PDB header: hydrolaseChain: A: PDB Molecule: luciferin regenerating enzyme;PDBTitle: luciferin-regenerating enzyme solved by siras using xfel (refined2 against native data)
43 c2qc5A_
not modelled
99.4
11
PDB header: lyaseChain: A: PDB Molecule: streptogramin b lactonase;PDBTitle: streptogramin b lyase structure
44 c3soqA_
not modelled
99.4
13
PDB header: protein binding/antagonistChain: A: PDB Molecule: low-density lipoprotein receptor-related protein 6;PDBTitle: the structure of the first ywtd beta propeller domain of lrp6 in2 complex with a dkk1 peptide
45 c5tzsT_
not modelled
99.4
16
PDB header: translationChain: T: PDB Molecule: utp21;PDBTitle: architecture of the yeast small subunit processome
46 c2j57J_
not modelled
99.4
14
PDB header: oxidoreductaseChain: J: PDB Molecule: methylamine dehydrogenase heavy chain;PDBTitle: x-ray reduced paraccocus denitrificans methylamine2 dehydrogenase n-quinol in complex with amicyanin.
47 d1pbyb_
not modelled
99.4
19
Fold: 7-bladed beta-propellerSuperfamily: YVTN repeat-like/Quinoprotein amine dehydrogenaseFamily: Quinohemoprotein amine dehydrogenase B chain
48 c2p9wA_
not modelled
99.3
21
PDB header: allergenChain: A: PDB Molecule: mal s 1 allergenic protein;PDBTitle: crystal structure of the major malassezia sympodialis allergen mala s2 1
49 c3g4hB_
not modelled
99.3
15
PDB header: hydrolaseChain: B: PDB Molecule: regucalcin;PDBTitle: crystal structure of human senescence marker protein-30 (zinc bound)
50 c4qrjA_
not modelled
99.3
14
PDB header: hydrolaseChain: A: PDB Molecule: putative 6-phosphogluconolactonase;PDBTitle: crystal structure of a putative 6-phosphogluconolactonase2 (bacuni_04672) from bacteroides uniformis atcc 8492 at 2.20 a3 resolution
51 c5i2tA_
not modelled
99.3
9
PDB header: biosynthetic proteinChain: A: PDB Molecule: periodic tryptophan protein 2;PDBTitle: domain characterization of the wd protein pwp2 and their relevance in2 ribosome biogenesis
52 d1ri6a_
not modelled
99.3
17
Fold: 7-bladed beta-propellerSuperfamily: Putative isomerase YbhEFamily: Putative isomerase YbhE
53 c3fgbB_
not modelled
99.3
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein q89zh8_bactn;PDBTitle: crystal structure of the q89zh8_bactn protein from bacteroides2 thetaiotaomicron. northeast structural genomics consortium target3 btr289b.
54 c3tc9B_
not modelled
99.3
14
PDB header: hydrolaseChain: B: PDB Molecule: hypothetical hydrolase;PDBTitle: crystal structure of an auxiliary nutrient binding protein (bt_3476)2 from bacteroides thetaiotaomicron vpi-5482 at 2.23 a resolution
55 d2ghsa1
not modelled
99.3
16
Fold: 6-bladed beta-propellerSuperfamily: Calcium-dependent phosphotriesteraseFamily: SGL-like
56 c2ghsA_
not modelled
99.3
16
PDB header: calcium-binding proteinChain: A: PDB Molecule: agr_c_1268p;PDBTitle: crystal structure of a calcium-binding protein, regucalcin2 (agr_c_1268) from agrobacterium tumefaciens str. c58 at 1.55 a3 resolution
57 c3dsmA_
not modelled
99.3
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein bacuni_02894;PDBTitle: crystal structure of the surface layer protein bacuni_02894 from2 bacteroides uniformis, northeast structural genomics consortium3 target btr193d.
58 c2i0tB_
not modelled
99.3
16
PDB header: oxidoreductaseChain: B: PDB Molecule: aromatic amine dehydrogenase;PDBTitle: crystal structure of phenylacetaldehyde derived r-2 carbinolamine adduct of aromatic amine dehydrogenase
59 c2h47F_
not modelled
99.3
17
PDB header: oxidoreductase/electron transportChain: F: PDB Molecule: aromatic amine dehydrogenase;PDBTitle: crystal structure of an electron transfer complex between2 aromatic amine dephydrogenase and azurin from alcaligenes3 faecalis (form 1)
60 d1ijqa1
not modelled
99.3
18
Fold: 6-bladed beta-propellerSuperfamily: YWTD domainFamily: YWTD domain
61 c3c75J_
not modelled
99.3
20
PDB header: oxidoreductaseChain: J: PDB Molecule: methylamine dehydrogenase heavy chain;PDBTitle: paracoccus versutus methylamine dehydrogenase in complex2 with amicyanin
62 c4nsxA_
not modelled
99.3
16
PDB header: protein bindingChain: A: PDB Molecule: u3 small nucleolar rna-associated protein 21;PDBTitle: crystal structure of the utp21 tandem wd domain
63 d1npea_
not modelled
99.3
18
Fold: 6-bladed beta-propellerSuperfamily: YWTD domainFamily: YWTD domain
64 c3dr2A_
not modelled
99.3
9
PDB header: hydrolaseChain: A: PDB Molecule: exported gluconolactonase;PDBTitle: structural and functional analyses of xc5397 from2 xanthomonas campestris: a gluconolactonase important in3 glucose secondary metabolic pathways
65 c2qe8B_
not modelled
99.3
11
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative hydrolase (ava_4197) from anabaena2 variabilis atcc 29413 at 1.35 a resolution
66 d2dg1a1
not modelled
99.3
21
Fold: 6-bladed beta-propellerSuperfamily: Calcium-dependent phosphotriesteraseFamily: SGL-like
67 d1v04a_
not modelled
99.3
20
Fold: 6-bladed beta-propellerSuperfamily: Calcium-dependent phosphotriesteraseFamily: Serum paraoxonase/arylesterase 1, PON1
68 d1rwia_
not modelled
99.3
31
Fold: 6-bladed beta-propellerSuperfamily: NHL repeatFamily: NHL repeat
69 d1nira2
not modelled
99.3
12
Fold: 8-bladed beta-propellerSuperfamily: C-terminal (heme d1) domain of cytochrome cd1-nitrite reductaseFamily: C-terminal (heme d1) domain of cytochrome cd1-nitrite reductase
70 c6ck1C_
not modelled
99.3
16
PDB header: metal binding proteinChain: C: PDB Molecule: a1b2f4 protein;PDBTitle: crystal structure of paracoccus denitrificans aztd
71 c6cmkB_
not modelled
99.3
14
PDB header: metal binding proteinChain: B: PDB Molecule: aztd protein;PDBTitle: crystal structure of citrobacter koseri aztd
72 c5a1vL_
not modelled
99.2
16
PDB header: transport proteinChain: L: PDB Molecule: coatomer subunit beta;PDBTitle: the structure of the copi coat linkage i
73 c6nd4T_
not modelled
99.2
16
PDB header: ribosomeChain: T: PDB Molecule: utp21;PDBTitle: conformational switches control early maturation of the eukaryotic2 small ribosomal subunit
74 c6d69A_
not modelled
99.2
19
PDB header: protein bindingChain: A: PDB Molecule: nhl repeat region of d. melanogaster thin;PDBTitle: crystal structure of the nhl repeat region of d. melanogaster thin
75 d2madh_
not modelled
99.2
11
Fold: 7-bladed beta-propellerSuperfamily: YVTN repeat-like/Quinoprotein amine dehydrogenaseFamily: Methylamine dehydrogenase, H-chain
76 c2fp8A_
not modelled
99.2
19
PDB header: lyaseChain: A: PDB Molecule: strictosidine synthase;PDBTitle: structure of strictosidine synthase, the biosynthetic entry to the2 monoterpenoid indole alkaloid family
77 c6nd4O_
not modelled
99.2
8
PDB header: ribosomeChain: O: PDB Molecule: utp1;PDBTitle: conformational switches control early maturation of the eukaryotic2 small ribosomal subunit
78 c5nzvC_
not modelled
99.2
17
PDB header: transport proteinChain: C: PDB Molecule: coatomer subunit beta';PDBTitle: the structure of the copi coat linkage iv
79 d2bbkh_
not modelled
99.2
24
Fold: 7-bladed beta-propellerSuperfamily: YVTN repeat-like/Quinoprotein amine dehydrogenaseFamily: Methylamine dehydrogenase, H-chain
80 c3iytG_
not modelled
99.2
8
PDB header: apoptosisChain: G: PDB Molecule: apoptotic protease-activating factor 1;PDBTitle: structure of an apoptosome-procaspase-9 card complex
81 c5a1uC_
not modelled
99.2
12
PDB header: transport proteinChain: C: PDB Molecule: coatomer subunit alpha;PDBTitle: the structure of the copi coat triad
82 c5a1vK_
not modelled
99.2
10
PDB header: transport proteinChain: K: PDB Molecule: coatomer subunit alpha;PDBTitle: the structure of the copi coat linkage i
83 c3mkqA_
not modelled
99.2
16
PDB header: transport proteinChain: A: PDB Molecule: coatomer beta'-subunit;PDBTitle: crystal structure of yeast alpha/betaprime-cop subcomplex of the copi2 vesicular coat
84 c3nokB_
not modelled
99.1
9
PDB header: transferaseChain: B: PDB Molecule: glutaminyl cyclase;PDBTitle: crystal structure of myxococcus xanthus glutaminyl cyclase
85 c3qqzA_
not modelled
99.1
9
PDB header: metal binding proteinChain: A: PDB Molecule: putative uncharacterized protein yjik;PDBTitle: crystal structure of the c-terminal domain of the yjik protein from2 escherichia coli cft073
86 c2ymuA_
not modelled
99.1
19
PDB header: unknown functionChain: A: PDB Molecule: wd-40 repeat protein;PDBTitle: structure of a highly repetitive propeller structure
87 d1q7fa_
not modelled
99.1
15
Fold: 6-bladed beta-propellerSuperfamily: NHL repeatFamily: NHL repeat
88 c3e5zA_
not modelled
99.1
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative gluconolactonase;PDBTitle: x-ray structure of the putative gluconolactonase in protein family2 pf08450. northeast structural genomics consortium target drr130.
89 c6igbB_
not modelled
99.1
16
PDB header: hydrolaseChain: B: PDB Molecule: periplasmic gluconolactonase, ppgl;PDBTitle: the structure of pseudomonas aeruginosa periplasmic gluconolactonase,2 ppgl
90 c5juyB_
not modelled
99.1
8
PDB header: apoptosisChain: B: PDB Molecule: apoptotic protease-activating factor 1;PDBTitle: active human apoptosome with procaspase-9
91 c3dm0A_
not modelled
99.1
9
PDB header: sugar binding protein,signaling proteinChain: A: PDB Molecule: maltose-binding periplasmic protein fused with rack1;PDBTitle: maltose binding protein fusion with rack1 from a. thaliana
92 c1ijqA_
not modelled
99.1
22
PDB header: lipid transportChain: A: PDB Molecule: low-density lipoprotein receptor;PDBTitle: crystal structure of the ldl receptor ywtd-egf domain pair
93 c6nd4Q_
not modelled
99.1
10
PDB header: ribosomeChain: Q: PDB Molecule: utp12;PDBTitle: conformational switches control early maturation of the eukaryotic2 small ribosomal subunit
94 c6fqlA_
not modelled
99.1
25
PDB header: rna binding proteinChain: A: PDB Molecule: e3 ubiquitin-protein ligase trim71;PDBTitle: crystal structure of danio rerio lin41 filamin-nhl domains in complex2 with mab-10 3'utr 13mer rna
95 c4wjsA_
not modelled
99.0
15
PDB header: biosynthetic proteinChain: A: PDB Molecule: rsa4;PDBTitle: crystal structure of rsa4 from chaetomium thermophilum
96 d1crua_
not modelled
99.0
19
Fold: 6-bladed beta-propellerSuperfamily: Soluble quinoprotein glucose dehydrogenaseFamily: Soluble quinoprotein glucose dehydrogenase
97 c4wjuB_
not modelled
99.0
15
PDB header: biosynthetic proteinChain: B: PDB Molecule: ribosome assembly protein 4;PDBTitle: crystal structure of rsa4 from saccharomyces cerevisiae
98 c1nr0A_
not modelled
99.0
13
PDB header: structural proteinChain: A: PDB Molecule: actin interacting protein 1;PDBTitle: two seven-bladed beta-propeller domains revealed by the2 structure of a c. elegans homologue of yeast actin3 interacting protein 1 (aip1).
99 c5thaA_
not modelled
99.0
15
PDB header: rna binding proteinChain: A: PDB Molecule: gem-associated protein 5;PDBTitle: gemin5 wd40 repeats in complex with a guanosyl moiety
100 c5cvoD_
not modelled
99.0
9
PDB header: hydrolase/protein bindingChain: D: PDB Molecule: wd repeat-containing protein 48;PDBTitle: wdr48:usp46~ubiquitin ternary complex
101 c2oajA_
not modelled
99.0
13
PDB header: endocytosis/exocytosisChain: A: PDB Molecule: protein sni1;PDBTitle: crystal structure of sro7 from s. cerevisiae
102 d1ospo_
not modelled
99.0
6
Fold: open-sided beta-meanderSuperfamily: Outer surface proteinFamily: Outer surface protein
103 c5ch2A_
not modelled
99.0
13
PDB header: transferaseChain: A: PDB Molecule: putative polycomb protein eed;PDBTitle: crystal structure of an active polycomb repressive complex 2 in the2 basal state
104 d1tl2a_
not modelled
98.9
14
Fold: 5-bladed beta-propellerSuperfamily: Tachylectin-2Family: Tachylectin-2
105 c5cvoA_
not modelled
98.9
10
PDB header: hydrolase/protein bindingChain: A: PDB Molecule: wd repeat-containing protein 48;PDBTitle: wdr48:usp46~ubiquitin ternary complex
106 c3a9gA_
not modelled
98.9
18
PDB header: oxidoreductaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of pqq-dependent sugar dehydrogenase apo-form
107 c5a5uB_
not modelled
98.9
10
PDB header: translationChain: B: PDB Molecule: eukaryotic translation initiation factor 3 subunit b;PDBTitle: structure of mammalian eif3 in the context of the 43s preinitiation2 complex
108 c2w18A_
not modelled
98.8
14
PDB header: nuclear proteinChain: A: PDB Molecule: partner and localizer of brca2;PDBTitle: crystal structure of the c-terminal wd40 domain of human2 palb2
109 c5k1bB_
not modelled
98.8
10
PDB header: protein binding/hydrolaseChain: B: PDB Molecule: wd repeat-containing protein 48;PDBTitle: crystal structure of the uaf1/usp12 complex in f222 space group
110 c6nd4S_
not modelled
98.8
13
PDB header: ribosomeChain: S: PDB Molecule: utp18;PDBTitle: conformational switches control early maturation of the eukaryotic2 small ribosomal subunit
111 c1n6dE_
not modelled
98.8
9
PDB header: hydrolaseChain: E: PDB Molecule: tricorn protease;PDBTitle: tricorn protease in complex with tetrapeptide chloromethyl2 ketone derivative
112 c1k32E_
not modelled
98.8
9
PDB header: hydrolaseChain: E: PDB Molecule: tricorn protease;PDBTitle: crystal structure of the tricorn protease
113 c4yczA_
not modelled
98.8
12
PDB header: structural proteinChain: A: PDB Molecule: fusion protein of sec13 and nup145c;PDBTitle: y-complex hub (nup85-nup120-nup145c-sec13 complex) from m. thermophila2 (a.k.a. t. heterothallica)
114 c4yhcA_
not modelled
98.8
12
PDB header: structural proteinChain: A: PDB Molecule: sterol regulatory element-binding protein cleavage-PDBTitle: crystal structure of the wd40 domain of scap from fission yeast
115 c2aq5A_
not modelled
98.8
16
PDB header: structural proteinChain: A: PDB Molecule: coronin-1a;PDBTitle: crystal structure of murine coronin-1
116 c1pi6A_
not modelled
98.8
10
PDB header: protein bindingChain: A: PDB Molecule: actin interacting protein 1;PDBTitle: yeast actin interacting protein 1 (aip1), orthorhombic crystal form
117 c6nd4H_
not modelled
98.8
11
PDB header: ribosomeChain: H: PDB Molecule: utp17;PDBTitle: conformational switches control early maturation of the eukaryotic2 small ribosomal subunit
118 c3j65q_
not modelled
98.7
15
PDB header: ribosomeChain: Q: PDB Molecule: 60s ribosomal protein l18;PDBTitle: arx1 pre-60s particle. this entry contains the r-proteins and2 biogenesis factors.
119 c5cvlA_
not modelled
98.7
10
PDB header: protein bindingChain: A: PDB Molecule: wd repeat-containing protein 48;PDBTitle: wdr48 (uaf-1), residues 2-580
120 c4xfvA_
not modelled
98.7
12
PDB header: translationChain: A: PDB Molecule: elongator complex protein 2;PDBTitle: crystal structure of elp2