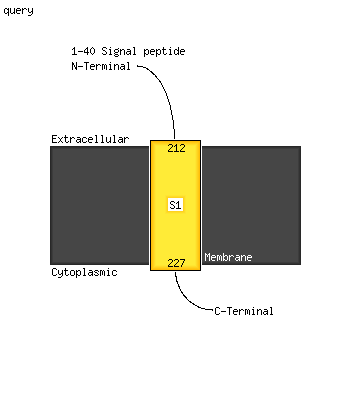
1 d2icha1
100.0
35
Fold: AttH-likeSuperfamily: AttH-likeFamily: AttH-like
2 d1tvna1
66.7
13
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
3 c2b4rQ_
62.4
24
PDB header: oxidoreductaseChain: Q: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 plasmodium falciparum at 2.25 angstrom resolution reveals intriguing3 extra electron density in the active site
4 d1egza_
54.6
13
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
5 c5fipA_
54.1
17
PDB header: hydrolaseChain: A: PDB Molecule: gh5 cellulase;PDBTitle: discovery and characterization of a novel thermostable and2 highly halotolerant gh5 cellulase from an icelandic hot3 spring isolate
6 d1whva_
50.2
23
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD
7 d1g01a_
50.1
16
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
8 c2i5pO_
47.3
20
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase 1;PDBTitle: crystal structure of glyceraldehyde-3-phosphate2 dehydrogenase isoform 1 from k. marxianus
9 c2d2iO_
44.9
35
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of nadp-dependent glyceraldehyde-3-2 phosphate dehydrogenase from synechococcus sp. complexed3 with nadp+
10 c3b20R_
43.9
35
PDB header: oxidoreductaseChain: R: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase (nadp+);PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase2 complexed with nadfrom synechococcus elongatus"
11 c3docD_
43.7
35
PDB header: oxidoreductaseChain: D: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of trka glyceraldehyde-3-phosphate dehydrogenase2 from brucella melitensis
12 c4dibF_
43.3
30
PDB header: oxidoreductaseChain: F: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: the crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 bacillus anthracis str. sterne
13 c1rm4O_
42.2
30
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase a;PDBTitle: crystal structure of recombinant photosynthetic glyceraldehyde-3-2 phosphate dehydrogenase a4 isoform, complexed with nadp
14 c2pkrI_
41.5
30
PDB header: oxidoreductaseChain: I: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase aor;PDBTitle: crystal structure of (a+cte)4 chimeric form of photosyntetic2 glyceraldehyde-3-phosphate dehydrogenase, complexed with nadp
15 c1hdgO_
41.1
40
PDB header: oxidoreductase (aldehy(d)-nad(a))Chain: O: PDB Molecule: holo-d-glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: the crystal structure of holo-glyceraldehyde-3-phosphate dehydrogenase2 from the hyperthermophilic bacterium thermotoga maritima at 2.53 angstroms resolution
16 c3h9eO_
39.9
30
PDB header: oxidoreductaseChain: O: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase, testis-specific;PDBTitle: crystal structure of human sperm-specific glyceraldehyde-3-phosphate2 dehydrogenase (gapds) complex with nad and phosphate
17 c2x5kO_
39.9
25
PDB header: oxidoreductaseChain: O: PDB Molecule: d-erythrose-4-phosphate dehydrogenase;PDBTitle: structure of an active site mutant of the d-erythrose-4-phosphate2 dehydrogenase from e. coli
18 c1s7cA_
38.7
25
PDB header: structural genomics, oxidoreductaseChain: A: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase a;PDBTitle: crystal structure of mes buffer bound form of glyceraldehyde 3-2 phosphate dehydrogenase from escherichia coli
19 c1ihxD_
38.4
25
PDB header: oxidoreductaseChain: D: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of two d-glyceraldehyde-3-phosphate2 dehydrogenase complexes: a case of asymmetry
20 c3hjaB_
38.3
30
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 borrelia burgdorferi
21 c6ok4A_
not modelled
36.7
30
PDB header: oxidoreductaseChain: A: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase (gapdh)2 from chlamydia trachomatis with bound nad
22 c3sthA_
not modelled
34.2
25
PDB header: oxidoreductaseChain: A: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 toxoplasma gondii
23 c2gd1P_
not modelled
31.2
35
PDB header: oxidoreductase(aldehyde(d)-nad(a))Chain: P: PDB Molecule: apo-d-glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: coenzyme-induced conformational changes in glyceraldehyde-3-2 phosphate dehydrogenase from bacillus stearothermophillus
24 c3cieC_
not modelled
27.5
42
PDB header: oxidoreductaseChain: C: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde 3-phosphate2 dehydrogenase from cryptosporidium parvum
25 c5sydA_
not modelled
24.1
15
PDB header: oxidoreductaseChain: A: PDB Molecule: azurin, chimeric construct;PDBTitle: circularly permutated azurin (cpaz) based on p. aeruginosa azurin2 sequence
26 c5ld5C_
not modelled
24.0
27
PDB header: oxidoreductaseChain: C: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of a bacterial dehydrogenase at 2.19 angstroms2 resolution
27 d1xnka1
not modelled
23.3
17
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Xylanase/endoglucanase 11/12
28 c2dvyA_
not modelled
22.3
26
PDB header: hydrolaseChain: A: PDB Molecule: restriction endonuclease pabi;PDBTitle: crystal structure of restriction endonucleases pabi
29 c2cksB_
not modelled
22.1
19
PDB header: hydrolaseChain: B: PDB Molecule: endoglucanase e-5;PDBTitle: x-ray crystal structure of the catalytic domain of thermobifida fusca2 endoglucanase cel5a (e5)
30 c5uhxA_
not modelled
21.1
18
PDB header: hydrolaseChain: A: PDB Molecule: carbohydrate-active enzyme;PDBTitle: structure of cellulase cel5c_1
31 d1ei5a1
not modelled
19.8
26
Fold: Streptavidin-likeSuperfamily: D-aminopeptidase, middle and C-terminal domainsFamily: D-aminopeptidase, middle and C-terminal domains
32 c4xzbA_
not modelled
19.8
17
PDB header: hydrolaseChain: A: PDB Molecule: cela;PDBTitle: endo-glucanase gscela p1
33 c2r5iL_
not modelled
19.1
19
PDB header: viral proteinChain: L: PDB Molecule: l1 protein;PDBTitle: pentamer structure of major capsid protein l1 of human papilloma virus2 type 18
34 c5jyfB_
not modelled
19.0
42
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: structures of streptococcus agalactiae gbs gapdh in different2 enzymatic states
35 d1r44a_
not modelled
18.8
56
Fold: Hedgehog/DD-peptidaseSuperfamily: Hedgehog/DD-peptidaseFamily: VanX-like
36 c5ihsA_
not modelled
17.8
22
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase, glycoside hydrolase family 5 protein;PDBTitle: structure of chu_2103 from cytophaga hutchinsonii
37 c4htyA_
not modelled
17.2
24
PDB header: hydrolaseChain: A: PDB Molecule: cellulase;PDBTitle: crystal structure of a metagenome-derived cellulase cel5a
38 c4hcgA_
not modelled
16.7
7
PDB header: oxidoreductaseChain: A: PDB Molecule: cupredoxin 1;PDBTitle: uncharacterized cupredoxin-like domain protein cupredoxin_1 with zinc2 bound from bacillus anthracis
39 c5ur0B_
not modelled
15.6
33
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystallographic structure of glyceraldehyde-3-phosphate dehydrogenase2 from naegleria gruberi
40 c4o2hB_
not modelled
15.1
41
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein bcam1869;PDBTitle: crystal structure of bcam1869 protein (rsam homolog) from burkholderia2 cenocepacia
41 c2ep7B_
not modelled
14.9
31
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: structural study of project id aq_1065 from aquifex aeolicus vf5
42 c2dcjA_
not modelled
14.6
22
PDB header: hydrolaseChain: A: PDB Molecule: xylanase j;PDBTitle: a two-domain structure of alkaliphilic xynj from bacillus sp. 41m-1
43 d2pyta1
not modelled
14.5
13
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: EutQ-like
44 c1cerC_
not modelled
14.5
33
PDB header: oxidoreductase (aldehyde(d)-nad(a))Chain: C: PDB Molecule: holo-d-glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: determinants of enzyme thermostability observed in the2 molecular structure of thermus aquaticus d-glyceraldehyde-3 3-phosphate dehydrogenase at 2.5 angstroms resolution
45 c3hq4R_
not modelled
14.4
35
PDB header: oxidoreductaseChain: R: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase 1;PDBTitle: crystal structure of c151s mutant of glyceraldehyde-3-phosphate2 dehydrogenase 1 (gapdh1) complexed with nad from staphylococcus3 aureus mrsa252 at 2.2 angstrom resolution
46 c1obfO_
not modelled
14.0
38
PDB header: glycolytic pathwayChain: O: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: the crystal structure of glyceraldehyde 3-phosphate2 dehydrogenase from alcaligenes xylosoxidans at 1.7 a3 resolution.
47 c5j9gB_
not modelled
13.4
41
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-p dehydrogenase;PDBTitle: structure of lactobacillus acidophilus glyceraldehyde-3-phosphate2 dehydrogenase at 2.21 angstrom resolution
48 c1i32D_
not modelled
12.8
33
PDB header: oxidoreductaseChain: D: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: leishmania mexicana glyceraldehyde-3-phosphate2 dehydrogenase in complex with inhibitors
49 d1wuba_
not modelled
11.8
18
Fold: Streptavidin-likeSuperfamily: YceI-likeFamily: YceI-like
50 d1y0ga_
not modelled
11.7
26
Fold: Streptavidin-likeSuperfamily: YceI-likeFamily: YceI-like
51 c4qx6A_
not modelled
11.6
30
PDB header: oxidoreductaseChain: A: PDB Molecule: glyceraldehyde 3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate dehydrogenase from2 streptococcus agalactiae nem316 at 2.46 angstrom resolution
52 c3ctrA_
not modelled
11.6
31
PDB header: hydrolaseChain: A: PDB Molecule: poly(a)-specific ribonuclease parn;PDBTitle: crystal structure of the rrm-domain of the poly(a)-specific2 ribonuclease parn bound to m7gtp
53 c5lhxA_
not modelled
11.6
22
PDB header: structural proteinChain: A: PDB Molecule: serine/threonine-protein kinase plk4;PDBTitle: pb3 domain of drosophila melanogaster plk4 (sak)
54 c3pzvB_
not modelled
11.4
17
PDB header: hydrolaseChain: B: PDB Molecule: endoglucanase;PDBTitle: c2 crystal form of the endo-1,4-beta-glucanase from bacillus subtilis2 168
55 c2r5kE_
not modelled
11.2
20
PDB header: viral proteinChain: E: PDB Molecule: major capsid protein l1;PDBTitle: pentamer structure of major capsid protein l1 of human papilloma virus2 type 11
56 d7a3ha_
not modelled
11.0
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
57 c5xf9F_
not modelled
11.0
8
PDB header: oxidoreductaseChain: F: PDB Molecule: nad-reducing hydrogenase;PDBTitle: crystal structure of nad+-reducing [nife]-hydrogenase in the air-2 oxidized state
58 d1cc3a_
not modelled
10.6
13
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like
59 c5ixhB_
not modelled
10.6
18
PDB header: unknown functionChain: B: PDB Molecule: ycei-like domain protein;PDBTitle: crystal structure of burkholderia cenocepacia bcna
60 c6f0kA_
not modelled
10.4
25
PDB header: membrane proteinChain: A: PDB Molecule: cytochrome c family protein;PDBTitle: alternative complex iii
61 c2aanA_
not modelled
10.1
14
PDB header: electron transportChain: A: PDB Molecule: auracyanin a;PDBTitle: auracyanin a: a "blue" copper protein from the green thermophilic2 photosynthetic bacterium,chloroflexus aurantiacus
62 c3hpeB_
not modelled
9.7
25
PDB header: transport proteinChain: B: PDB Molecule: conserved hypothetical secreted protein;PDBTitle: crystal structure of ycei (hp1286) from helicobacter pylori
63 c5i2uB_
not modelled
9.3
20
PDB header: hydrolaseChain: B: PDB Molecule: cellulase;PDBTitle: crystal structure of a novel halo-tolerant cellulase from soil2 metagenome
64 c2z5cA_
not modelled
9.3
18
PDB header: chaperone/hydrolaseChain: A: PDB Molecule: protein ypl144w;PDBTitle: crystal structure of a novel chaperone complex for yeast 20s2 proteasome assembly
65 c2fgsA_
not modelled
9.3
17
PDB header: lipid binding proteinChain: A: PDB Molecule: putative periplasmic protein;PDBTitle: crystal structure of campylobacter jejuni ycei protein,2 structural genomics
66 c3q34A_
not modelled
9.3
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ycei-like family protein;PDBTitle: the crystal structure of ycei-like family protein from pseudomonas2 syringae
67 d1ecea_
not modelled
9.0
15
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
68 d1plqa2
not modelled
8.9
14
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase processivity factor
69 c4f2eA_
not modelled
8.4
10
PDB header: metal transportChain: A: PDB Molecule: cupa;PDBTitle: crystal structure of the streptococcus pneumoniae d39 copper chaperone2 cupa with cu(i)
70 c3bvdC_
not modelled
8.4
59
PDB header: oxidoreductaseChain: C: PDB Molecule: cytochrome c oxidase polypeptide 2a;PDBTitle: structure of surface-engineered cytochrome ba3 oxidase from thermus2 thermophilus under xenon pressure, 100psi 5min
71 d1xmec1
not modelled
8.4
59
Fold: Single transmembrane helixSuperfamily: Bacterial ba3 type cytochrome c oxidase subunit IIaFamily: Bacterial ba3 type cytochrome c oxidase subunit IIa
72 c5ixgD_
not modelled
8.3
17
PDB header: unknown functionChain: D: PDB Molecule: ycei;PDBTitle: crystal structure of burkholderia cenocepacia bcnb
73 c6amgA_
not modelled
8.2
18
PDB header: metal binding proteinChain: A: PDB Molecule: cytochrome p460;PDBTitle: cyt p460 of nitrosomonas sp. al212
74 d1miua4
not modelled
8.0
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB
75 c4bjrA_
not modelled
7.9
40
PDB header: ligaseChain: A: PDB Molecule: pup--protein ligase, prokaryotic ubiquitin-like proteinPDBTitle: crystal structure of the complex between prokaryotic2 ubiquitin-like protein pup and its ligase pafa
76 c2h47C_
not modelled
7.9
13
PDB header: oxidoreductase/electron transportChain: C: PDB Molecule: azurin;PDBTitle: crystal structure of an electron transfer complex between2 aromatic amine dephydrogenase and azurin from alcaligenes3 faecalis (form 1)
77 c2mqcA_
not modelled
7.8
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: nmr structure of the protein bvu_0925 from bacteroides vulgatus atcc2 8482
78 d1ajoa_
not modelled
7.7
38
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Glycosyl hydrolases family 16
79 c4y66C_
not modelled
7.6
18
PDB header: cell cycleChain: C: PDB Molecule: mnd1;PDBTitle: crystal structure of giardia lamblia hop2-mnd1 complex
80 c4g6tB_
not modelled
7.5
18
PDB header: chaperoneChain: B: PDB Molecule: type iii effector hopa1;PDBTitle: structure of the hopa1-scha chaperone-effector complex
81 c1kjkA_
not modelled
7.3
7
PDB header: viral proteinChain: A: PDB Molecule: integrase;PDBTitle: solution structure of the lambda integrase amino-terminal2 domain
82 d1z1ba1
not modelled
7.3
7
Fold: DNA-binding domainSuperfamily: DNA-binding domainFamily: lambda integrase N-terminal domain
83 c2x34A_
not modelled
6.7
11
PDB header: carbohydrate-binding proteinChain: A: PDB Molecule: cellulose-binding protein, x158;PDBTitle: structure of a polyisoprenoid binding domain from saccharophagus2 degradans implicated in plant cell wall breakdown
84 c4nohA_
not modelled
6.7
19
PDB header: lipid binding proteinChain: A: PDB Molecule: lipoprotein, putative;PDBTitle: 1.5 angstrom crystal structure of putative lipoprotein from bacillus2 anthracis.
85 c3iyjE_
not modelled
6.6
20
PDB header: virusChain: E: PDB Molecule: major capsid protein l1;PDBTitle: bovine papillomavirus type 1 outer capsid
86 c2l9uB_
not modelled
6.4
30
PDB header: membrane proteinChain: B: PDB Molecule: receptor tyrosine-protein kinase erbb-3;PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
87 c2l9uA_
not modelled
6.4
30
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-3;PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
88 c1jauA_
not modelled
6.3
50
PDB header: viral proteinChain: A: PDB Molecule: transmembrane glycoprotein (gp41);PDBTitle: nmr solution structure of the trp-rich peptide of hiv gp412 bound to dpc micelles
89 c4fd7A_
not modelled
6.3
19
PDB header: transferaseChain: A: PDB Molecule: putative arylalkylamine n-acetyltransferase 7;PDBTitle: crystal structure of insect putative arylalkylamine n-2 acetyltransferase 7 from the yellow fever mosquito aedes aegypt
90 c4kmaA_
not modelled
6.3
41
PDB header: protein bindingChain: A: PDB Molecule: gm14141p;PDBTitle: crystal structure of drosophila suppressor of fused
91 c6igdG_
not modelled
6.3
19
PDB header: structural proteinChain: G: PDB Molecule: major capsid protein l1;PDBTitle: crystal structure of hpv58/33 chimeric l1 pentamer
92 c3pm6B_
not modelled
6.2
18
PDB header: lyaseChain: B: PDB Molecule: putative fructose-bisphosphate aldolase;PDBTitle: crystal structure of a putative fructose-1,6-biphosphate aldolase from2 coccidioides immitis solved by combined sad mr
93 d2c0ha1
not modelled
6.2
23
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
94 d1dzla_
not modelled
6.1
20
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Group I dsDNA virusesFamily: Papovaviridae-like VP
95 c1dzlA_
not modelled
6.1
20
PDB header: virusChain: A: PDB Molecule: late major capsid protein l1;PDBTitle: l1 protein of human papillomavirus 16
96 d2diga1
not modelled
6.0
23
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: Tudor domain
97 c5b0uB_
not modelled
6.0
28
PDB header: luminescent proteinChain: B: PDB Molecule: oplophorus-luciferin 2-monooxygenase catalytic subunit;PDBTitle: crystal structure of the mutated 19 kda protein of oplophorus2 luciferase (nanokaz)
98 d1cuoa_
not modelled
5.9
16
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like
99 d1m1la_
not modelled
5.9
41
Fold: Suppressor of Fused, N-terminal domainSuperfamily: Suppressor of Fused, N-terminal domainFamily: Suppressor of Fused, N-terminal domain