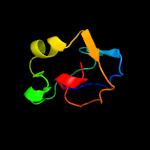
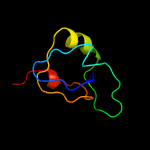
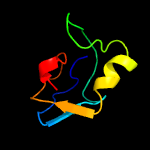
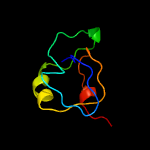
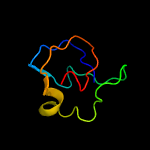
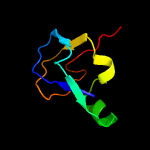
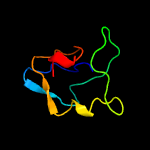
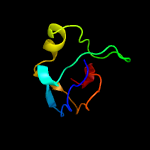
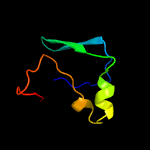
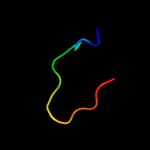
1 c3teeA_
99.4
20
PDB header: chaperoneChain: A: PDB Molecule: flagella basal body p-ring formation protein flga;PDBTitle: crystal structure of salmonella flga in open form
2 c3frnA_
99.2
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: flagellar protein flga;PDBTitle: crystal structure of flagellar protein flga from thermotoga maritima2 msb8
3 d2zdra1
98.0
20
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain
4 d1vlia1
97.8
6
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain
5 c1xuzA_
97.7
20
PDB header: biosynthetic proteinChain: A: PDB Molecule: polysialic acid capsule biosynthesis protein siac;PDBTitle: crystal structure analysis of sialic acid synthase (neub)from2 neisseria meningitidis, bound to mn2+, phosphoenolpyruvate, and n-3 acetyl mannosaminitol
6 c3g8rA_
97.4
19
PDB header: biosynthetic proteinChain: A: PDB Molecule: probable spore coat polysaccharide biosynthesis protein e;PDBTitle: crystal structure of putative spore coat polysaccharide biosynthesis2 protein e from chromobacterium violaceum atcc 12472
7 c1vliA_
97.4
8
PDB header: biosynthetic proteinChain: A: PDB Molecule: spore coat polysaccharide biosynthesis protein spse;PDBTitle: crystal structure of spore coat polysaccharide biosynthesis protein2 spse (bsu37870) from bacillus subtilis at 2.38 a resolution
8 d1opsa_
96.6
27
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain
9 d1c8aa2
96.2
25
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain
10 c1wvoA_
95.7
20
PDB header: transferaseChain: A: PDB Molecule: sialic acid synthase;PDBTitle: solution structure of rsgi ruh-029, an antifreeze protein2 like domain in human n-acetylneuraminic acid phosphate3 synthase gene.
11 d3nlaa_
95.7
25
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain
12 c1c8aA_
94.9
25
PDB header: antifreeze proteinChain: A: PDB Molecule: protein (antifreeze protein type iii);PDBTitle: nmr structure of intramolecular dimer antifreeze protein2 rd3, 40 sa structures
13 c3upyB_
92.1
20
PDB header: oxidoreductaseChain: B: PDB Molecule: oxidoreductase;PDBTitle: crystal structure of the brucella abortus enzyme catalyzing the first2 committed step of the methylerythritol 4-phosphate pathway.
14 d1ucsa_
85.0
23
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain
15 d1hg7a_
81.6
25
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain
16 c3k3sG_
72.0
21
PDB header: hydrolaseChain: G: PDB Molecule: altronate hydrolase;PDBTitle: crystal structure of altronate hydrolase (fragment 1-84) from shigella2 flexneri.
17 c6chgC_
57.2
33
PDB header: transferaseChain: C: PDB Molecule: histone-lysine n-methyltransferase, h3 lysine-4 specific;PDBTitle: crystal structure of the yeast compass catalytic module
18 c5ht6B_
57.0
27
PDB header: transferaseChain: B: PDB Molecule: histone-lysine n-methyltransferase 2e;PDBTitle: crystal structure of the set domain of the human mll52 methyltransferase
19 c4z4pA_
51.3
20
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase 2d;PDBTitle: structure of the mll4 set domain
20 d2g46a1
51.1
13
Fold: beta-clipSuperfamily: SET domainFamily: Viral histone H3 Lysine 27 Methyltransferase
21 c3f9xA_
not modelled
50.9
27
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase setd8;PDBTitle: structural insights into lysine multiple methylation by set domain2 methyltransferases, set8-y334f / h4-lys20me2 / adohcy
22 d2f69a2
not modelled
49.0
13
Fold: beta-clipSuperfamily: SET domainFamily: Histone lysine methyltransferases
23 c2w5zA_
not modelled
47.9
33
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase hrx;PDBTitle: ternary complex of the mixed lineage leukaemia (mll1) set2 domain with the cofactor product s-adenosylhomocysteine3 and histone peptide.
24 d1h3ia2
not modelled
44.2
13
Fold: beta-clipSuperfamily: SET domainFamily: Histone lysine methyltransferases
25 c3opeA_
not modelled
42.9
40
PDB header: transferaseChain: A: PDB Molecule: probable histone-lysine n-methyltransferase ash1l;PDBTitle: structural basis of auto-inhibitory mechanism of histone2 methyltransferase
26 c6bx3E_
not modelled
42.3
33
PDB header: gene regulation/transferaseChain: E: PDB Molecule: histone-lysine n-methyltransferase, h3 lysine-4 specific;PDBTitle: structure of histone h3k4 methyltransferase
27 c3h6lA_
not modelled
42.2
20
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase setd2;PDBTitle: methyltransferase domain of human set domain-containing protein 2
28 c6agoA_
not modelled
42.0
40
PDB header: transferase/gene regulationChain: A: PDB Molecule: histone-lysine n-methyltransferase ash1l;PDBTitle: crystal structure of mrg15-ash1l histone methyltransferase complex
29 c4o30B_
not modelled
41.7
33
PDB header: transferaseChain: B: PDB Molecule: histone-lysine n-methyltransferase atxr6, putative;PDBTitle: crystal structure of atxr5 in complex with histone h3.1 and adohcy
30 c5h6zA_
not modelled
41.1
7
PDB header: transcriptionChain: A: PDB Molecule: set domain-containing protein 7;PDBTitle: crystal structure of set7, a novel histone methyltransferase in2 schizossacharomyces pombe
31 c4mi0A_
not modelled
39.6
20
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase ezh2;PDBTitle: human enhancer of zeste (drosophila) homolog 2(ezh2)
32 c4rz0B_
not modelled
39.5
13
PDB header: transferaseChain: B: PDB Molecule: pfl0690c;PDBTitle: crystal structure of plasmodium falciparum putative histone2 methyltransferase pfl0690c
33 c3bo5A_
not modelled
39.0
33
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase setmar;PDBTitle: crystal structure of methyltransferase domain of human histone-lysine2 n-methyltransferase setmar
34 c3ooiA_
not modelled
38.6
40
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase, h3 lysine-36 and h4PDBTitle: crystal structure of human histone-lysine n-methyltransferase nsd1 set2 domain in complex with s-adenosyl-l-methionine
35 c2r3aA_
not modelled
38.2
20
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase suv39h2;PDBTitle: methyltransferase domain of human suppressor of variegation2 3-9 homolog 2
36 d1ml9a_
not modelled
37.6
40
Fold: beta-clipSuperfamily: SET domainFamily: Histone lysine methyltransferases
37 c3hnaA_
not modelled
36.1
13
PDB header: hydrolase/hydrolase regulatorChain: A: PDB Molecule: histone-lysine n-methyltransferase, h3 lysine-9 specific 5;PDBTitle: crystal structure of catalytic domain of human euchromatic histone2 methyltransferase 1 in complex with sah and mono-methylated h3k93 peptide
38 c2o8jC_
not modelled
34.4
27
PDB header: transferaseChain: C: PDB Molecule: histone-lysine n-methyltransferase, h3 lysine-9PDBTitle: human euchromatic histone methyltransferase 2
39 c1xqhE_
not modelled
34.3
13
PDB header: transferaseChain: E: PDB Molecule: histone-lysine n-methyltransferase, h3 lysine-4PDBTitle: crystal structure of a ternary complex of the2 methyltransferase set9 (also known as set7/9) with a p533 peptide and sah
40 c3n71A_
not modelled
31.5
8
PDB header: transcriptionChain: A: PDB Molecule: histone lysine methyltransferase smyd1;PDBTitle: crystal structure of cardiac specific histone methyltransferase smyd1
41 d1mvha_
not modelled
31.2
33
Fold: beta-clipSuperfamily: SET domainFamily: Histone lysine methyltransferases
42 c1mvhA_
not modelled
31.2
33
PDB header: transferaseChain: A: PDB Molecule: cryptic loci regulator 4;PDBTitle: structure of the set domain histone lysine2 methyltransferase clr4
43 c3lazB_
not modelled
31.1
7
PDB header: lyaseChain: B: PDB Molecule: d-galactarate dehydratase;PDBTitle: the crystal structure of the n-terminal domain of d-2 galactarate dehydratase from escherichia coli cft073
44 c4ldgA_
not modelled
29.7
27
PDB header: unknown functionChain: A: PDB Molecule: protein with a set domain within carboxy region;PDBTitle: crystal structure of cpset8 from cryptosporidium, cgd4_370
45 c2qpwA_
not modelled
28.8
20
PDB header: transcriptionChain: A: PDB Molecule: pr domain zinc finger protein 2;PDBTitle: methyltransferase domain of human pr domain-containing2 protein 2
46 c3s8pA_
not modelled
28.3
33
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase suv420h1;PDBTitle: crystal structure of the set domain of human histone-lysine n-2 methyltransferase suv420h1 in complex with s-adenosyl-l-methionine
47 c6a5mA_
not modelled
27.0
20
PDB header: gene regulationChain: A: PDB Molecule: histone-lysine n-methyltransferase, h3 lysine-9 specificPDBTitle: crystal structure of arabidopsis thaliana suvh6 in complex with sam,2 form 2
48 c4ytlB_
not modelled
26.1
16
PDB header: transcriptionChain: B: PDB Molecule: transcription elongation factor spt5;PDBTitle: structure of the kow2-kow3 domain of transcription elongation factor2 spt5.
49 c3rayA_
not modelled
25.9
40
PDB header: transcriptionChain: A: PDB Molecule: pr domain-containing protein 11;PDBTitle: crystal structure of methyltransferase domain of human pr domain-2 containing protein 11
50 c3rq4A_
not modelled
25.7
33
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase suv420h2;PDBTitle: crystal structure of suppressor of variegation 4-20 homolog 2
51 c1h3iB_
not modelled
23.4
13
PDB header: transferaseChain: B: PDB Molecule: histone h3 lysine 4 specific methyltransferase;PDBTitle: crystal structure of the histone methyltransferase set7/9
52 c4nj5A_
not modelled
23.1
33
PDB header: metal binding proteinChain: A: PDB Molecule: probable histone-lysine n-methyltransferase, h3 lysine-9PDBTitle: crystal structure of suvh9
53 c3ep0A_
not modelled
22.1
21
PDB header: transferaseChain: A: PDB Molecule: pr domain zinc finger protein 12;PDBTitle: methyltransferase domain of human pr domain-containing2 protein 12
54 c3qwvA_
not modelled
21.8
23
PDB header: transferaseChain: A: PDB Molecule: set and mynd domain-containing protein 2;PDBTitle: crystal structure of histone lysine methyltransferase smyd2 in complex2 with the cofactor product adohcy
55 c3mekA_
not modelled
19.0
46
PDB header: transferaseChain: A: PDB Molecule: set and mynd domain-containing protein 3;PDBTitle: crystal structure of human histone-lysine n-methyltransferase smyd3 in2 complex with s-adenosyl-l-methionine
56 d2h2ja2
not modelled
18.9
7
Fold: beta-clipSuperfamily: SET domainFamily: RuBisCo LSMT catalytic domain
57 c6fndC_
not modelled
17.0
33
PDB header: transferaseChain: C: PDB Molecule: apical complex lysine methyltransferase;PDBTitle: crystal structure of toxoplasma gondii akmt
58 c3oxgA_
not modelled
16.0
46
PDB header: transferaseChain: A: PDB Molecule: set and mynd domain-containing protein 3;PDBTitle: human lysine methyltransferase smyd3 in complex with adohcy (form iii)
59 c1wzoC_
not modelled
15.9
23
PDB header: isomeraseChain: C: PDB Molecule: hpce;PDBTitle: crystal structure of the hpce from thermus thermophilus hb8
60 d1nkqa_
not modelled
14.8
16
Fold: FAHSuperfamily: FAHFamily: FAH
61 c4qeoA_
not modelled
14.3
40
PDB header: transcription/dnaChain: A: PDB Molecule: histone-lysine n-methyltransferase, h3 lysine-9 specificPDBTitle: crystal structure of kryptonite in complex with mchh dna, h3(1-15)2 peptide and sah
62 c4e5sC_
not modelled
14.1
27
PDB header: hydrolaseChain: C: PDB Molecule: mccflike protein (ba_5613);PDBTitle: crystal structure of mccflike protein (ba_5613) from bacillus2 anthracis str. ames
63 d1gtta2
not modelled
14.1
20
Fold: FAHSuperfamily: FAHFamily: FAH
64 c3db5A_
not modelled
13.8
23
PDB header: transferaseChain: A: PDB Molecule: pr domain zinc finger protein 4;PDBTitle: crystal structure of methyltransferase domain of human pr domain-2 containing protein 4
65 c6jvwA_
not modelled
13.3
18
PDB header: hydrolaseChain: A: PDB Molecule: maleylpyruvate hydrolase;PDBTitle: crystal structure of maleylpyruvate hydrolase from sphingobium sp.2 syk-6 in complex with manganese (ii) ion and pyruvate
66 d1sawa_
not modelled
12.4
12
Fold: FAHSuperfamily: FAHFamily: FAH
67 d1hh2p1
not modelled
12.2
19
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
68 c2hj1A_
not modelled
12.1
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a 3d domain-swapped dimer of protein hi0395 from2 haemophilus influenzae
69 d2hj1a1
not modelled
12.1
20
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: HI0395-like
70 c3gjzB_
not modelled
12.0
20
PDB header: immune systemChain: B: PDB Molecule: microcin immunity protein mccf;PDBTitle: crystal structure of microcin immunity protein mccf from bacillus2 anthracis str. ames
71 c5zz5D_
not modelled
11.6
18
PDB header: gene regulationChain: D: PDB Molecule: redox-sensing transcriptional repressor rex;PDBTitle: redox-sensing transcriptional repressor rex
72 d2do3a1
not modelled
11.6
13
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: SPT5 KOW domain-like
73 c6iymB_
not modelled
11.5
20
PDB header: hydrolaseChain: B: PDB Molecule: 5-oxopent-3-ene-1,2,5-tricarboxylate decarboxylase;PDBTitle: fumarylacetoacetate hydrolase (eafah) from psychrophilic2 exiguobacterium antarcticum
74 c5ij7B_
not modelled
11.4
20
PDB header: transferase/transferase inhibitorChain: B: PDB Molecule: enhancer of zeste homolog 2 (ezh2),histone-lysine n-PDBTitle: structure of hs/acprc2 in complex with a pyridone inhibitor
75 c3qdfA_
not modelled
11.4
19
PDB header: isomeraseChain: A: PDB Molecule: 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase;PDBTitle: crystal structure of 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase2 from mycobacterium marinum
76 c2l55A_
not modelled
11.3
13
PDB header: metal binding proteinChain: A: PDB Molecule: silb,silver efflux protein, mfp component of the threePDBTitle: solution structure of the c-terminal domain of silb from cupriavidus2 metallidurans
77 c5f1yA_
not modelled
11.1
21
PDB header: hydrolaseChain: A: PDB Molecule: mccc family protein;PDBTitle: crystal structure of ba3275, the member of s66 family of serine2 peptidases
78 c3tlgB_
not modelled
10.7
27
PDB header: hydrolaseChain: B: PDB Molecule: mccf;PDBTitle: microcin c7 self immunity protein mccf in the inactive mutant apo2 state
79 c4h1hB_
not modelled
10.5
18
PDB header: hydrolaseChain: B: PDB Molecule: lmo1638 protein;PDBTitle: crystal structure of mccf homolog from listeria monocytogenes egd-e
80 d1e0ta3
not modelled
9.8
24
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: PK C-terminal domain-likeFamily: Pyruvate kinase, C-terminal domain
81 c6f0kA_
not modelled
9.1
14
PDB header: membrane proteinChain: A: PDB Molecule: cytochrome c family protein;PDBTitle: alternative complex iii
82 c4maqB_
not modelled
9.0
16
PDB header: hydrolaseChain: B: PDB Molecule: putative fumarylpyruvate hydrolase;PDBTitle: crystal structure of a putative fumarylpyruvate hydrolase from2 burkholderia cenocepacia
83 c5wg6C_
not modelled
9.0
20
PDB header: transferase/transferase inhibitorChain: C: PDB Molecule: histone-lysine n-methyltransferase ezh2,polycomb proteinPDBTitle: human polycomb repressive complex 2 in complex with gsk126 inhibitor
84 c3qxyA_
not modelled
8.6
31
PDB header: transferaseChain: A: PDB Molecule: n-lysine methyltransferase setd6;PDBTitle: human setd6 in complex with rela lys310
85 c2e6zA_
not modelled
8.2
31
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt5;PDBTitle: solution structure of the second kow motif of human2 transcription elongation factor spt5
86 c1zrsB_
not modelled
8.1
18
PDB header: hydrolaseChain: B: PDB Molecule: hypothetical protein;PDBTitle: wild-type ld-carboxypeptidase
87 c4dbhA_
not modelled
8.0
14
PDB header: isomeraseChain: A: PDB Molecule: 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase;PDBTitle: crystal structure of cg1458 with inhibitor
88 d2g50a3
not modelled
7.8
16
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: PK C-terminal domain-likeFamily: Pyruvate kinase, C-terminal domain
89 c1ri9A_
not modelled
7.5
19
PDB header: signaling proteinChain: A: PDB Molecule: fyn-binding protein;PDBTitle: structure of a helically extended sh3 domain of the t cell2 adapter protein adap
90 d1ri9a_
not modelled
7.5
19
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain
91 c1p0yA_
not modelled
7.5
23
PDB header: transferaseChain: A: PDB Molecule: ribulose-1,5 bisphosphate carboxylase/oxygenasePDBTitle: crystal structure of the set domain of lsmt bound to2 melysine and adohcy
92 d1hr0w_
not modelled
7.3
26
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
93 c4hkrB_
not modelled
7.2
9
PDB header: transport proteinChain: B: PDB Molecule: calcium release-activated calcium channel protein 1;PDBTitle: calcium release-activated calcium (crac) channel orai
94 d1ah9a_
not modelled
6.9
32
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
95 d2auna2
not modelled
6.8
12
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: LD-carboxypeptidase A N-terminal domain-like
96 c6flqF_
not modelled
6.5
22
PDB header: transcriptionChain: F: PDB Molecule: transcription termination/antitermination protein nusa;PDBTitle: cryoem structure of e.coli rna polymerase paused elongation complex2 bound to nusa
97 c3zr6A_
not modelled
6.5
30
PDB header: hydrolaseChain: A: PDB Molecule: galactocerebrosidase;PDBTitle: structure of galactocerebrosidase from mouse in complex with galactose
98 c4ql5A_
not modelled
6.4
26
PDB header: translationChain: A: PDB Molecule: translation initiation factor if-1;PDBTitle: crystal structure of translation initiation factor if-1 from2 streptococcus pneumoniae tigr4
99 c3i4oA_
not modelled
6.1
26
PDB header: translationChain: A: PDB Molecule: translation initiation factor if-1;PDBTitle: crystal structure of translation initiation factor 1 from2 mycobacterium tuberculosis