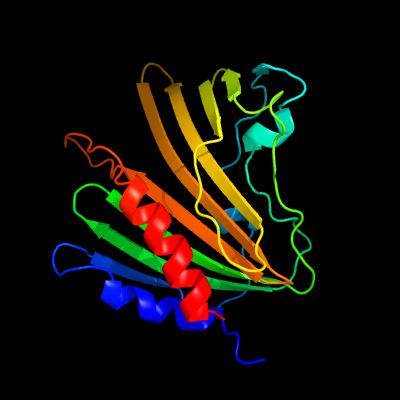
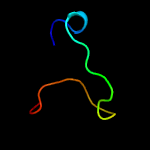
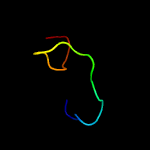
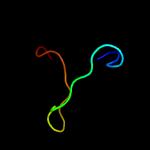
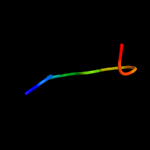
1 c4tmdA_
100.0
58
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: x-ray structure of putative uncharacterized protein (rv0999 ortholog)2 from mycobacterium smegmatis
2 c4h69A_
49.6
33
PDB header: isomeraseChain: A: PDB Molecule: allene oxide cyclase;PDBTitle: crystal structure of the allene oxide cyclase 2 from physcomitrella2 patens complexed with substrate analog
3 d1zvca1
48.2
29
Fold: AOC barrel-likeSuperfamily: Allene oxide cyclase-likeFamily: Allene oxide cyclase-like
4 d2brja1
25.7
25
Fold: AOC barrel-likeSuperfamily: Allene oxide cyclase-likeFamily: Allene oxide cyclase-like
5 c2dzqA_
17.0
30
PDB header: transcriptionChain: A: PDB Molecule: general transcription factor ii-i repeat domain-PDBTitle: solution structure of rsgi ruh-066, a gtf2i domain in human2 cdna
6 c2ed2A_
16.4
17
PDB header: transcriptionChain: A: PDB Molecule: general transcription factor ii-i;PDBTitle: solution structure of rsgi ruh-069, a gtf2i domain in human2 cdna
7 c2dn4A_
16.2
17
PDB header: transcriptionChain: A: PDB Molecule: general transcription factor ii-i;PDBTitle: solution structure of rsgi ruh-060, a gtf2i domain in human2 cdna
8 c2d99A_
16.2
30
PDB header: transcriptionChain: A: PDB Molecule: general transcription factor ii-i repeat domain-PDBTitle: solution structure of rsgi ruh-048, a gtf2i domain in human2 cdna
9 d1q60a_
16.2
26
Fold: GTF2I-like repeatSuperfamily: GTF2I-like repeatFamily: GTF2I-like repeat
10 c2dn5A_
15.4
26
PDB header: transcriptionChain: A: PDB Molecule: general transcription factor ii-i repeat domain-PDBTitle: solution structure of rsgi ruh-057, a gtf2i domain in human2 cdna
11 c2ejeA_
14.8
30
PDB header: transcriptionChain: A: PDB Molecule: general transcription factor ii-i;PDBTitle: solution structure of rsgi ruh-071, a gtf2i domain in human2 cdna
12 c1af7A_
14.4
33
PDB header: methyltransferaseChain: A: PDB Molecule: chemotaxis receptor methyltransferase cher;PDBTitle: cher from salmonella typhimurium
13 c2e3lA_
14.1
22
PDB header: transcriptionChain: A: PDB Molecule: transcription factor gtf2ird2 beta;PDBTitle: solution structure of rsgi ruh-068, a gtf2i domain in human2 cdna
14 c2dzrA_
14.1
26
PDB header: transcriptionChain: A: PDB Molecule: general transcription factor ii-i repeat domain-PDBTitle: solution structure of rsgi ruh-067, a gtf2i domain in human2 cdna
15 c5xlyA_
12.7
56
PDB header: transferaseChain: A: PDB Molecule: chemotaxis protein methyltransferase 1;PDBTitle: crystal structure of cher1 in complex with c-di-gmp-bound mapz
16 c5mz61_
12.1
27
PDB header: cell cycleChain: 1: PDB Molecule: separase;PDBTitle: cryo-em structure of a separase-securin complex from caenorhabditis2 elegans at 3.8 a resolution
17 c5u1sA_
11.3
50
PDB header: hydrolaseChain: A: PDB Molecule: separin;PDBTitle: crystal structure of the saccharomyces cerevisiae separase-securin2 complex at 3.0 angstrom resolution
18 c2m9lA_
10.6
67
PDB header: toxinChain: A: PDB Molecule: beta-theraphotoxin-tp1a;PDBTitle: solution structure of protoxin-1
19 c2yuvA_
10.5
11
PDB header: cell adhesionChain: A: PDB Molecule: myosin-binding protein c, slow-type;PDBTitle: solution structure of 2nd immunoglobulin domain of slow2 type myosin-binding protein c
20 c6nu9A_
9.7
22
PDB header: viral proteinChain: A: PDB Molecule: zinc-binding non-structural protein;PDBTitle: crystal structure of a zinc-binding non-structural protein from the2 hepatitis e virus
21 d1wmhb_
not modelled
9.7
25
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: PB1 domain
22 c3ld1A_
not modelled
8.7
21
PDB header: hydrolaseChain: A: PDB Molecule: replicase polyprotein 1a;PDBTitle: crystal structure of ibv nsp2a
23 c5ftwA_
not modelled
8.4
50
PDB header: transferaseChain: A: PDB Molecule: chemotaxis protein methyltransferase;PDBTitle: crystal structure of glutamate o-methyltransferase in2 complex with s- adenosyl-l-homocysteine (sah) from3 bacillus subtilis
24 c3nuhB_
not modelled
7.7
32
PDB header: isomeraseChain: B: PDB Molecule: dna gyrase subunit b;PDBTitle: a domain insertion in e. coli gyrb adopts a novel fold that plays a2 critical role in gyrase function
25 c2fvnA_
not modelled
7.3
24
PDB header: cell adhesionChain: A: PDB Molecule: protein afad;PDBTitle: the fibrillar tip complex of the afa/dr adhesins from2 pathogen e. coli displays synergistic binding to 5 1 and v3 3 integrins
26 c3zrhA_
not modelled
7.3
32
PDB header: hydrolaseChain: A: PDB Molecule: ubiquitin thioesterase zranb1;PDBTitle: crystal structure of the lys29, lys33-linkage-specific trabid otu2 deubiquitinase domain reveals an ankyrin-repeat ubiquitin binding3 domain (ankubd)
27 c4phxE_
not modelled
7.1
22
PDB header: cell adhesionChain: E: PDB Molecule: protein aggb;PDBTitle: crystal structure of aggb, the minor subunit of aggregative adherence2 fimbriae type i from the escherichia coli o4h104
28 c2ehdB_
not modelled
6.5
6
PDB header: oxidoreductaseChain: B: PDB Molecule: oxidoreductase, short-chain dehydrogenase/reductase family;PDBTitle: crystal structure analysis of oxidoreductase
29 c5fc2A_
not modelled
6.0
63
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: pamk, peptide containing a phospho-serine;PDBTitle: structure of a separase in complex with a pamk peptide containing a2 phospho-serine
30 c4bm5A_
not modelled
5.9
28
PDB header: protein transportChain: A: PDB Molecule: similar to chloroplast inner membrane protein tic110;PDBTitle: chloroplast inner membrane protein tic110
31 c2myxA_
not modelled
5.9
12
PDB header: ubiquitin-binding proteinChain: A: PDB Molecule: coupling of ubiquitin conjugation to er degradation proteinPDBTitle: structure of the cue domain of yeast cue1
32 c5sujB_
not modelled
5.8
19
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein lpg2148 from legionella2 pneumophila
33 c2jf1A_
not modelled
5.7
25
PDB header: cell adhesionChain: A: PDB Molecule: filamin-a;PDBTitle: crystal structure of the filamin a repeat 21 complexed with2 the integrin beta2 cytoplasmic tail peptide
34 d1te1b_
not modelled
5.3
27
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Xylanase/endoglucanase 11/12
35 c2vulA_
not modelled
5.2
24
PDB header: hydrolaseChain: A: PDB Molecule: gh11 xylanase;PDBTitle: thermostable mutant of environmentally isolated gh112 xylanase