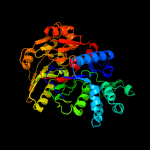
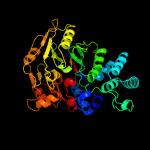
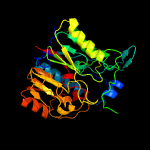
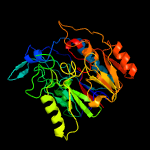
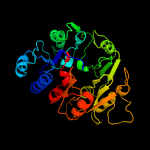
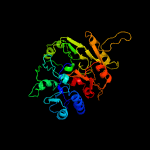
1 d1rxxa_
100.0
39
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Arginine deiminase
2 c4bofA_
100.0
38
PDB header: hydrolaseChain: A: PDB Molecule: arginine deiminase;PDBTitle: crystal structure of arginine deiminase from group a streptococcus
3 d1s9ra_
100.0
30
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Arginine deiminase
4 c4e4jJ_
100.0
30
PDB header: hydrolaseChain: J: PDB Molecule: arginine deiminase;PDBTitle: crystal structure of arginine deiminase from mycoplasma penetrans
5 c5wpiB_
100.0
16
PDB header: transferaseChain: B: PDB Molecule: hsva;PDBTitle: the virulence-associated protein hsva from the fire blight pathogen2 erwinia amylovora is a polyamine amidinotransferase
6 d1bwda_
100.0
18
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Amidinotransferase
7 c8jdwA_
100.0
18
PDB header: transferaseChain: A: PDB Molecule: protein (l-arginine:glycine amidinotransferase);PDBTitle: crystal structure of human l-arginine:glycine amidinotransferase in2 complex with l-alanine
8 c1jdwA_
100.0
17
PDB header: transferaseChain: A: PDB Molecule: l-arginine\:glycine amidinotransferase;PDBTitle: crystal structure and mechanism of l-arginine: glycine2 amidinotransferase: a mitochondrial enzyme involved in3 creatine biosynthesis
9 d1jdwa_
100.0
17
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Amidinotransferase
10 c3i4aA_
100.0
24
PDB header: hydrolaseChain: A: PDB Molecule: n(g),n(g)-dimethylarginine dimethylaminohydrolase 1;PDBTitle: crystal structure of dimethylarginine dimethylaminohydrolase-1 (ddah-2 1) in complex with n5-(1-iminopropyl)-l-ornithine
11 c2ci6A_
100.0
23
PDB header: hydrolaseChain: A: PDB Molecule: ng, ng-dimethylarginine dimethylaminohydrolase 1;PDBTitle: crystal structure of dimethylarginine2 dimethylaminohydrolase i bound with zinc low ph
12 d1h70a_
100.0
23
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Dimethylarginine dimethylaminohydrolase DDAH
13 c6b2wB_
99.6
13
PDB header: hydrolaseChain: B: PDB Molecule: putative peptidyl-arginine deiminase family protein;PDBTitle: c. jejuni c315s agmatine deiminase with substrate bound
14 d2ewoa1
99.6
18
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Porphyromonas-type peptidylarginine deiminase
15 d1xkna_
99.6
19
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Porphyromonas-type peptidylarginine deiminase
16 d1zbra1
99.5
19
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Porphyromonas-type peptidylarginine deiminase
17 c2jerG_
99.5
21
PDB header: hydrolaseChain: G: PDB Molecule: agmatine deiminase;PDBTitle: agmatine deiminase of enterococcus faecalis catalyzing its2 reaction.
18 d2jera1
99.5
19
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Porphyromonas-type peptidylarginine deiminase
19 d1vkpa_
99.5
18
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Porphyromonas-type peptidylarginine deiminase
20 d2cmua1
99.5
14
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Porphyromonas-type peptidylarginine deiminase
21 c4ytgA_
not modelled
99.4
12
PDB header: hydrolaseChain: A: PDB Molecule: peptidylarginine deiminase;PDBTitle: crystal structure of porphyromonas gingivalis peptidylarginine2 deiminase (ppad) mutant c351a in complex with dipeptide met-arg.
22 d2dexx3
not modelled
94.2
12
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Peptidylarginine deiminase Pad4, catalytic C-terminal domain
23 c4n2gA_
not modelled
94.0
17
PDB header: hydrolaseChain: A: PDB Molecule: protein-arginine deiminase type-2;PDBTitle: crystal structure of protein arginine deiminase 2 (d169a, 10 mm ca2+)
24 c4n2kA_
not modelled
93.5
17
PDB header: hydrolaseChain: A: PDB Molecule: protein-arginine deiminase type-2;PDBTitle: crystal structure of protein arginine deiminase 2 (q350a, 0 mm ca2+)
25 c5hp5A_
not modelled
93.4
13
PDB header: hydrolaseChain: A: PDB Molecule: protein-arginine deiminase type-1;PDBTitle: srtucture of human peptidylarginine deiminase type i (pad1)
26 c2dexX_
not modelled
92.7
13
PDB header: hydrolaseChain: X: PDB Molecule: protein-arginine deiminase type iv;PDBTitle: crystal structure of human peptidylarginine deiminase 4 in complex2 with histone h3 n-terminal peptide including arg17
27 d2ebfx2
not modelled
46.3
17
Fold: EreA/ChaN-likeSuperfamily: EreA/ChaN-likeFamily: PMT domain-like
28 c6rdu2_
not modelled
43.8
13
PDB header: proton transportChain: 2: PDB Molecule: asa-2: polytomella f-atp synthase associated subunit 2;PDBTitle: cryo-em structure of polytomella f-atp synthase, rotary substate 1e,2 monomer-masked refinement
29 c4o9uB_
not modelled
42.9
46
PDB header: membrane proteinChain: B: PDB Molecule: nad(p) transhydrogenase subunit beta;PDBTitle: mechanism of transhydrogenase coupling proton translocation and2 hydride transfer
30 d1d4oa_
not modelled
40.7
21
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Transhydrogenase domain III (dIII)
31 d1pnoa_
not modelled
40.7
38
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Transhydrogenase domain III (dIII)
32 c4rrfD_
not modelled
39.6
22
PDB header: ligaseChain: D: PDB Molecule: threonine--trna ligase;PDBTitle: editing domain of threonyl-trna synthetase from methanococcus2 jannaschii with l-ser3aa
33 c1pt9B_
not modelled
38.9
21
PDB header: oxidoreductaseChain: B: PDB Molecule: nad(p) transhydrogenase, mitochondrial;PDBTitle: crystal structure analysis of the diii component of transhydrogenase2 with a thio-nicotinamide nucleotide analogue
34 c2bruC_
not modelled
37.9
38
PDB header: oxidoreductaseChain: C: PDB Molecule: nad(p) transhydrogenase subunit beta;PDBTitle: complex of the domain i and domain iii of escherichia coli2 transhydrogenase
35 c2hl2A_
not modelled
37.7
25
PDB header: ligaseChain: A: PDB Molecule: threonyl-trna synthetase;PDBTitle: crystal structure of the editing domain of threonyl-trna2 synthetase from pyrococcus abyssi in complex with an3 analog of seryladenylate
36 d1iuka_
not modelled
37.1
16
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain
37 c4k2bA_
not modelled
32.5
15
PDB header: transferaseChain: A: PDB Molecule: ntd biosynthesis operon protein ntda;PDBTitle: crystal structure of ntda from bacillus subtilis in complex with the2 internal aldimine
38 c3zs7A_
not modelled
30.1
24
PDB header: transferaseChain: A: PDB Molecule: pyridoxal kinase;PDBTitle: crystal structure of pyridoxal kinase from trypanosoma brucei
39 d1daaa_
not modelled
29.5
11
Fold: D-aminoacid aminotransferase-like PLP-dependent enzymesSuperfamily: D-aminoacid aminotransferase-like PLP-dependent enzymesFamily: D-aminoacid aminotransferase-like PLP-dependent enzymes
40 c3ff4A_
not modelled
26.9
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein chu_1412
41 c4ovkA_
not modelled
26.7
24
PDB header: solute-binding proteinChain: A: PDB Molecule: periplasmic binding protein;PDBTitle: crystal structure of periplasmic solute binding protein from2 veillonella parvula dsm 2008
42 c1xtzA_
not modelled
26.4
22
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase;PDBTitle: crystal structure of the s. cerevisiae d-ribose-5-phosphate isomerase:2 comparison with the archeal and bacterial enzymes
43 c4lhdB_
not modelled
24.6
15
PDB header: oxidoreductaseChain: B: PDB Molecule: glycine dehydrogenase [decarboxylating];PDBTitle: crystal structure of synechocystis sp. pcc 6803 glycine decarboxylase2 (p-protein), holo form with pyridoxal-5'-phosphate and glycine,3 closed flexible loop
44 d1u6ma_
not modelled
23.9
10
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT
45 d1ynha1
not modelled
23.5
20
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Succinylarginine dihydrolase-like
46 d2itba1
not modelled
23.4
25
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: MiaE-like
47 c3u7jA_
not modelled
23.1
35
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose-5-phosphate isomerase a from burkholderia2 thailandensis
48 c4inpA_
not modelled
22.5
24
PDB header: transport proteinChain: A: PDB Molecule: iron (iii) abc transporter, periplasmic iron-bindingPDBTitle: the crystal structure of helicobacter pylori ceue (hp1561) with ni(ii)2 bound
49 c4hn9B_
not modelled
22.4
24
PDB header: transport proteinChain: B: PDB Molecule: iron complex transport system substrate-binding protein;PDBTitle: crystal structure of iron abc transporter solute-binding protein from2 eubacterium eligens
50 c4x84C_
not modelled
21.5
30
PDB header: isomeraseChain: C: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose-5-phosphate isomerase a from pseudomonas2 aeruginosa
51 c4yajA_
not modelled
21.4
23
PDB header: ligaseChain: A: PDB Molecule: alpha subunit of acetyl-coenzyme a synthetasePDBTitle: ca. korarchaeum cryptofilum dinucleotide forming acetyl-coenzyme a2 synthetase 1 (apo form)
52 c4ce5A_
not modelled
20.9
18
PDB header: transferaseChain: A: PDB Molecule: at-omegata;PDBTitle: first crystal structure of an (r)-selective omega-transaminase2 from aspergillus terreus
53 c5ux1D_
not modelled
20.5
28
PDB header: lyaseChain: D: PDB Molecule: trna-(ms(2)io(6)a)-hydroxylase-like;PDBTitle: protein 43 with aldehyde deformylating oxygenase activity from2 synechococcus
54 c4nmlA_
not modelled
19.4
32
PDB header: isomeraseChain: A: PDB Molecule: ribulose 5-phosphate isomerase;PDBTitle: 2.60 angstrom resolution crystal structure of putative ribose 5-2 phosphate isomerase from toxoplasma gondii me49 in complex with dl-3 malic acid
55 c3wwjE_
not modelled
19.3
23
PDB header: transferaseChain: E: PDB Molecule: (r)-amine transaminase;PDBTitle: crystal structure of an engineered sitagliptin-producing transaminase,2 ata-117-rd11
56 c4rrcA_
not modelled
18.1
17
PDB header: ligaseChain: A: PDB Molecule: probable threonine--trna ligase 2;PDBTitle: n-terminal editing domain of threonyl-trna synthetase from aeropyrum2 pernix with l-thr3aa (snapshot 3)
57 c2e58D_
not modelled
15.4
29
PDB header: transferaseChain: D: PDB Molecule: mnmc2;PDBTitle: crystal structure of mnmc2 from aquifex aeolicus
58 c5cm0A_
not modelled
14.6
14
PDB header: transferaseChain: A: PDB Molecule: branched-chain transaminase;PDBTitle: crystal structure of branched-chain aminotransferase from thermophilic2 archaea geoglobus acetivorans
59 c5dlcC_
not modelled
14.6
26
PDB header: transferaseChain: C: PDB Molecule: pyridoxine 5'-phosphate synthase;PDBTitle: x-ray crystal structure of a pyridoxine 5-prime-phosphate synthase2 from pseudomonas aeruginosa
60 c6eddB_
not modelled
14.2
10
PDB header: transferaseChain: B: PDB Molecule: acetyltransferase pa3944;PDBTitle: crystal structure of a gnat superfamily pa3944 acetyltransferase in2 complex with coa (p1 space group)
61 d1qwga_
not modelled
13.6
25
Fold: TIM beta/alpha-barrelSuperfamily: (2r)-phospho-3-sulfolactate synthase ComAFamily: (2r)-phospho-3-sulfolactate synthase ComA
62 d2aeea1
not modelled
13.4
22
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)
63 c4d6yA_
not modelled
13.4
24
PDB header: signaling proteinChain: A: PDB Molecule: bacterial regulatory, fis family protein;PDBTitle: crystal structure of the receiver domain of ntrx from2 brucella abortus in complex with beryllofluoride and3 magnesium
64 c1r0lD_
not modelled
13.3
15
PDB header: oxidoreductaseChain: D: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: 1-deoxy-d-xylulose 5-phosphate reductoisomerase from zymomonas mobilis2 in complex with nadph
65 c5ix3A_
not modelled
13.2
6
PDB header: transferaseChain: A: PDB Molecule: diamine n-acetyltransferase;PDBTitle: crystal structure of n-acetyltransferase from staphylococcus aureus.
66 d1m5wa_
not modelled
13.2
24
Fold: TIM beta/alpha-barrelSuperfamily: Pyridoxine 5'-phosphate synthaseFamily: Pyridoxine 5'-phosphate synthase
67 c2z14A_
not modelled
13.1
16
PDB header: signaling proteinChain: A: PDB Molecule: ef-hand domain-containing family member c2;PDBTitle: crystal structure of the n-terminal duf1126 in human ef-2 hand domain containing 2 protein
68 d1f52a1
not modelled
12.8
11
Fold: beta-Grasp (ubiquitin-like)Superfamily: Glutamine synthetase, N-terminal domainFamily: Glutamine synthetase, N-terminal domain
69 c5b6aA_
not modelled
12.7
18
PDB header: transferaseChain: A: PDB Molecule: pyridoxal kinase pdxy;PDBTitle: structure of pyridoxal kinasefrom pseudomonas aeruginosa
70 c1wv9B_
not modelled
12.3
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: rhodanese homolog tt1651;PDBTitle: crystal structure of rhodanese homolog tt1651 from an2 extremely thermophilic bacterium thermus thermophilus hb8
71 d1liua2
not modelled
12.3
19
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Pyruvate kinase
72 d1xhfa1
not modelled
12.1
14
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related
73 d1jkea_
not modelled
11.9
21
Fold: DTD-likeSuperfamily: DTD-likeFamily: DTD-like
74 c1nl0G_
not modelled
11.6
58
PDB header: immune systemChain: G: PDB Molecule: factor ix;PDBTitle: crystal structure of human factor ix gla domain in complex2 of an inhibitory antibody, 10c12
75 d1gr0a2
not modelled
11.5
31
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like
76 d1tjna_
not modelled
11.4
17
Fold: Chelatase-likeSuperfamily: ChelataseFamily: CbiX-like
77 c1tjnA_
not modelled
11.4
17
PDB header: lyaseChain: A: PDB Molecule: sirohydrochlorin cobaltochelatase;PDBTitle: crystal structure of hypothetical protein af0721 from archaeoglobus2 fulgidus
78 c3u0gA_
not modelled
11.1
18
PDB header: transferaseChain: A: PDB Molecule: putative branched-chain amino acid aminotransferase ilve;PDBTitle: crystal structure of branched-chain amino acid aminotransferase from2 burkholderia pseudomallei
79 c4s1hA_
not modelled
11.0
15
PDB header: transferaseChain: A: PDB Molecule: pyridoxal kinase;PDBTitle: pyridoxal kinase of entamoeba histolytica with adp
80 c3a14B_
not modelled
10.9
27
PDB header: oxidoreductaseChain: B: PDB Molecule: 1-deoxy-d-xylulose 5-phosphate reductoisomerase;PDBTitle: crystal structure of dxr from thermotoga maritima, in complex with2 nadph
81 c5uidC_
not modelled
10.8
18
PDB header: transferaseChain: C: PDB Molecule: aminotransferase tlmj;PDBTitle: the crystal structure of an aminotransferase tlmj from2 streptoalloteichus hindustanus
82 c2zu8A_
not modelled
10.6
19
PDB header: transferaseChain: A: PDB Molecule: mannosyl-3-phosphoglycerate synthase;PDBTitle: crystal structure of mannosyl-3-phosphoglycerate synthase2 from pyrococcus horikoshii
83 c2x4bA_
not modelled
10.4
17
PDB header: hydrolaseChain: A: PDB Molecule: limit dextrinase;PDBTitle: barley limit dextrinase in complex with beta-cyclodextrin
84 c4m0jA_
not modelled
10.3
18
PDB header: transferaseChain: A: PDB Molecule: d-amino acid aminotransferase;PDBTitle: crystal structure of a d-amino acid aminotransferase from burkholderia2 thailandensis e264
85 d1dcfa_
not modelled
10.2
31
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: Receiver domain of the ethylene receptor
86 c4rkmK_
not modelled
10.1
24
PDB header: unknown functionChain: K: PDB Molecule: mcca;PDBTitle: wolinella succinogenes octaheme sulfite reductase mcca, form i
87 c2rdmB_
not modelled
10.0
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: response regulator receiver protein;PDBTitle: crystal structure of response regulator receiver protein from2 sinorhizobium medicae wsm419
88 c3qb8A_
not modelled
9.9
12
PDB header: transferaseChain: A: PDB Molecule: a654l protein;PDBTitle: paramecium chlorella bursaria virus1 putative orf a654l is a polyamine2 acetyltransferase
89 c5joqA_
not modelled
9.6
14
PDB header: hydrolaseChain: A: PDB Molecule: lmo2184 protein;PDBTitle: crystal structure of an abc transporter substrate-binding protein from2 listeria monocytogenes egd-e
90 d1p0sl2
not modelled
9.6
58
Fold: GLA-domainSuperfamily: GLA-domainFamily: GLA-domain
91 d1zh2a1
not modelled
9.5
18
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related
92 c3otrC_
not modelled
9.5
16
PDB header: lyaseChain: C: PDB Molecule: enolase;PDBTitle: 2.75 angstrom crystal structure of enolase 1 from toxoplasma gondii
93 c5khlB_
not modelled
9.4
24
PDB header: transport proteinChain: B: PDB Molecule: hemin abc transporter, periplasmic hemin-binding proteinPDBTitle: crystal structure of periplasmic heme binding protein hutb of vibrio2 cholerae
94 d2fiaa1
not modelled
9.3
7
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT
95 d1gmua2
not modelled
9.3
23
Fold: Ferredoxin-likeSuperfamily: Urease metallochaperone UreE, C-terminal domainFamily: Urease metallochaperone UreE, C-terminal domain
96 c2wnsB_
not modelled
9.0
20
PDB header: transferaseChain: B: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: human orotate phosphoribosyltransferase (oprtase) domain of2 uridine 5'-monophosphate synthase (umps) in complex with3 its substrate orotidine 5'-monophosphate (omp)
97 c3hdoB_
not modelled
8.9
14
PDB header: transferaseChain: B: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: crystal structure of a histidinol-phosphate aminotransferase from2 geobacter metallireducens
98 d1xlqa1
not modelled
8.8
17
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related
99 c4p9eA_
not modelled
8.8
30
PDB header: hydrolaseChain: A: PDB Molecule: deoxycytidylate deaminase;PDBTitle: crystal structure of dcmp deaminase from the cyanophage s-tim5 in apo2 form