| 1 | c5hw4C_
|
|
 |
100.0 |
40 |
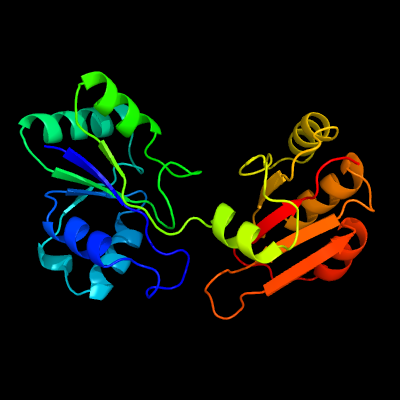
PDB header:transferase
Chain: C: PDB Molecule:ribosomal rna small subunit methyltransferase i;
PDBTitle: crystal structure of escherichia coli 16s rrna methyltransferase rsmi2 in complex with adomet
|
|
|
|
| 2 | c3kwpA_
|
|
 |
100.0 |
44 |
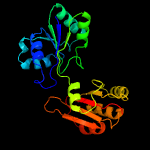
PDB header:transferase
Chain: A: PDB Molecule:predicted methyltransferase;
PDBTitle: crystal structure of putative methyltransferase from lactobacillus2 brevis
|
|
|
|
| 3 | d1cbfa_
|
|
 |
100.0 |
19 |
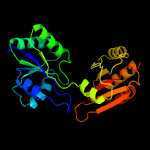
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
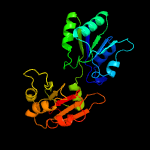
| 4 | c1cbfA_
|
|
 |
100.0 |
19 |
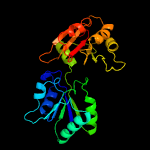
PDB header:methyltransferase
Chain: A: PDB Molecule:cobalt-precorrin-4 transmethylase;
PDBTitle: the x-ray structure of a cobalamin biosynthetic enzyme, cobalt2 precorrin-4 methyltransferase, cbif
|
|
|
|
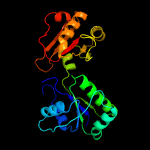
| 5 | d1wyza1
|
|
 |
100.0 |
21 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
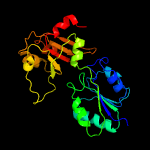
| 6 | c1pjtB_
|
|
 |
100.0 |
19 |
PDB header:transferase/oxidoreductase/lyase
Chain: B: PDB Molecule:siroheme synthase;
PDBTitle: the structure of the ser128ala point-mutant variant of cysg, the2 multifunctional methyltransferase/dehydrogenase/ferrochelatase for3 siroheme synthesis
|
|
|
|
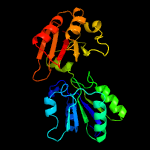
| 7 | c2yboA_
|
|
 |
100.0 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase;
PDBTitle: the x-ray structure of the sam-dependent uroporphyrinogen2 iii methyltransferase nire from pseudomonas aeruginosa in3 complex with sah
|
|
|
|
| 8 | c4e16A_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-4 c(11)-methyltransferase;
PDBTitle: precorrin-4 c(11)-methyltransferase from clostridium difficile
|
|
|
|
| 9 | d1s4da_
|
|
 |
100.0 |
17 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 10 | c3ndcB_
|
|
 |
100.0 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:precorrin-4 c(11)-methyltransferase;
PDBTitle: crystal structure of precorrin-4 c11-methyltransferase from2 rhodobacter capsulatus
|
|
|
|
| 11 | d1pjqa2
|
|
 |
100.0 |
19 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 12 | d1wdea_
|
|
 |
100.0 |
23 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 13 | c2zvbA_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-3 c17-methyltransferase;
PDBTitle: crystal structure of tt0207 from thermus thermophilus hb8
|
|
|
|
| 14 | c3nutC_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: C: PDB Molecule:precorrin-3 methylase;
PDBTitle: crystal structure of the methyltransferase cobj
|
|
|
|
| 15 | d1va0a1
|
|
 |
100.0 |
17 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
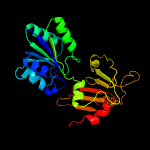
| 16 | d1ve2a1
|
|
 |
100.0 |
19 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
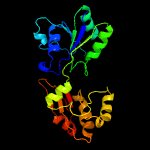
| 17 | c2qbuA_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-2 methyltransferase;
PDBTitle: crystal structure of methanothermobacter thermautotrophicus cbil
|
|
|
|
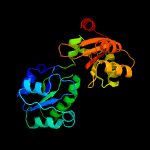
| 18 | d1vhva_
|
|
 |
100.0 |
18 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
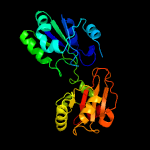
| 19 | d2deka1
|
|
 |
100.0 |
16 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 20 | c3nd1B_
|
|
 |
100.0 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:precorrin-6a synthase/cobf protein;
PDBTitle: crystal structure of precorrin-6a synthase from rhodobacter capsulatus
|
|
|
|
| 21 | c3i4tA_ |
|
not modelled |
100.0 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:diphthine synthase;
PDBTitle: crystal structure of putative diphthine synthase from entamoeba2 histolytica
|
|
|
| 22 | c2e0kA_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-2 c20-methyltransferase;
PDBTitle: crystal structure of cbil, a methyltransferase involved in anaerobic2 vitamin b12 biosynthesis
|
|
|
| 23 | c3fq6A_ |
|
not modelled |
100.0 |
51 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase;
PDBTitle: the crystal structure of a methyltransferase domain from bacteroides2 thetaiotaomicron vpi
|
|
|
| 24 | c2npnA_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:putative cobalamin synthesis related protein;
PDBTitle: crystal structure of putative cobalamin synthesis related protein2 (cobf) from corynebacterium diphtheriae
|
|
|
| 25 | c2bb3B_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:cobalamin biosynthesis precorrin-6y methylase (cbie);
PDBTitle: crystal structure of cobalamin biosynthesis precorrin-6y methylase2 (cbie) from archaeoglobus fulgidus
|
|
|
| 26 | d2bb3a1 |
|
not modelled |
100.0 |
19 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
| 27 | c5n0sA_ |
|
not modelled |
100.0 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:peptide n-methyltransferase;
PDBTitle: crystal structure of opha-deltac6 mutant y98a in complex with sam
|
|
|
| 28 | c3hh1D_ |
|
not modelled |
99.9 |
39 |
PDB header:transferase
Chain: D: PDB Molecule:tetrapyrrole methylase family protein;
PDBTitle: the structure of a tetrapyrrole methylase family protein domain from2 chlorobium tepidum tls
|
|
|
| 29 | c3q3vA_ |
|
not modelled |
80.6 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoglycerate kinase;
PDBTitle: crystal structure of phosphoglycerate kinase from campylobacter2 jejuni.
|
|
|
| 30 | c2px0D_ |
|
not modelled |
79.3 |
11 |
PDB header:biosynthetic protein
Chain: D: PDB Molecule:flagellar biosynthesis protein flhf;
PDBTitle: crystal structure of flhf complexed with gmppnp/mg(2+)
|
|
|
| 31 | d1phpa_ |
|
not modelled |
79.1 |
19 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
|
|
| 32 | d1vpea_ |
|
not modelled |
79.0 |
17 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
|
|
| 33 | c5b04G_ |
|
not modelled |
62.2 |
22 |
PDB header:translation
Chain: G: PDB Molecule:probable translation initiation factor eif-2b subunit
PDBTitle: crystal structure of the eukaryotic translation initiation factor 2b2 from schizosaccharomyces pombe
|
|
|
| 34 | d2ezla_ |
|
not modelled |
61.3 |
29 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Recombinase DNA-binding domain |
|
|
| 35 | c4qmkB_ |
|
not modelled |
60.5 |
38 |
PDB header:toxin
Chain: B: PDB Molecule:type iii secretion system effector protein exou;
PDBTitle: crystal structure of type iii effector protein exou (exou)
|
|
|
| 36 | c4akfA_ |
|
not modelled |
59.2 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:vipd;
PDBTitle: crystal structure of vipd from legionella pneumophila
|
|
|
| 37 | c5fyaA_ |
|
not modelled |
59.1 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:patatin-like protein, plpd;
PDBTitle: cubic crystal of the native plpd
|
|
|
| 38 | c3tu3B_ |
|
not modelled |
58.5 |
31 |
PDB header:toxin/toxin chaperone
Chain: B: PDB Molecule:exou;
PDBTitle: 1.92 angstrom resolution crystal structure of the full-length spcu in2 complex with full-length exou from the type iii secretion system of3 pseudomonas aeruginosa
|
|
|
| 39 | c4dg5A_ |
|
not modelled |
55.4 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoglycerate kinase;
PDBTitle: crystal structure of staphylococcal phosphoglycerate kinase
|
|
|
| 40 | d1v6sa_ |
|
not modelled |
51.7 |
18 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
|
|
| 41 | c1keeH_ |
|
not modelled |
51.1 |
19 |
PDB header:ligase
Chain: H: PDB Molecule:carbamoyl-phosphate synthetase small chain;
PDBTitle: inactivation of the amidotransferase activity of carbamoyl phosphate2 synthetase by the antibiotic acivicin
|
|
|
| 42 | c4ltyD_ |
|
not modelled |
50.4 |
27 |
PDB header:hydrolase
Chain: D: PDB Molecule:exonuclease subunit sbcd;
PDBTitle: crystal structure of e.coli sbcd at 1.8 a resolution
|
|
|
| 43 | c5kinC_ |
|
not modelled |
49.7 |
15 |
PDB header:lyase
Chain: C: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha beta complex from2 streptococcus pneumoniae
|
|
|
| 44 | d1xfia_ |
|
not modelled |
49.6 |
30 |
Fold:AF1104-like
Superfamily:AF1104-like
Family:AF1104-like |
|
|
| 45 | d1ybha1 |
|
not modelled |
49.4 |
9 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
|
|
| 46 | c3bioB_ |
|
not modelled |
46.9 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of oxidoreductase (gfo/idh/moca family member) from2 porphyromonas gingivalis w83
|
|
|
| 47 | c4akxB_ |
|
not modelled |
45.5 |
31 |
PDB header:transport protein
Chain: B: PDB Molecule:exou;
PDBTitle: structure of the heterodimeric complex exou-spcu from the type iii2 secretion system (t3ss) of pseudomonas aeruginosa
|
|
|
| 48 | c3navB_ |
|
not modelled |
45.2 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
|
|
| 49 | c5dboA_ |
|
not modelled |
45.2 |
28 |
PDB header:translation
Chain: A: PDB Molecule:translation initiation factor eif-2b-like protein;
PDBTitle: crystal structure of the tetrameric eif2b-beta2-delta2 complex from c.2 thermophilum
|
|
|
| 50 | c4da9C_ |
|
not modelled |
43.5 |
21 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:short-chain dehydrogenase/reductase;
PDBTitle: crystal structure of putative short-chain dehydrogenase/reductase from2 sinorhizobium meliloti 1021
|
|
|
| 51 | c6cluC_ |
|
not modelled |
43.2 |
17 |
PDB header:antimicrobial protein
Chain: C: PDB Molecule:dihydropteroate synthase;
PDBTitle: staphylococcus aureus dihydropteroate synthase (sadhps) f17l e208k2 double mutant structure
|
|
|
| 52 | c1cr6A_ |
|
not modelled |
40.6 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:epoxide hydrolase;
PDBTitle: crystal structure of murine soluble epoxide hydrolase2 complexed with cpu inhibitor
|
|
|
| 53 | c2cunA_ |
|
not modelled |
39.4 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoglycerate kinase;
PDBTitle: crystal structure of phosphoglycerate kinase from pyrococcus2 horikoshii ot3
|
|
|
| 54 | d2d59a1 |
|
not modelled |
38.7 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
| 55 | d1p3y1_ |
|
not modelled |
38.5 |
8 |
Fold:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Superfamily:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Family:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD |
|
|
| 56 | c2z2uA_ |
|
not modelled |
37.8 |
14 |
PDB header:metal binding protein
Chain: A: PDB Molecule:upf0026 protein mj0257;
PDBTitle: crystal structure of archaeal tyw1
|
|
|
| 57 | c4njkA_ |
|
not modelled |
37.2 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:7-carboxy-7-deazaguanine synthase;
PDBTitle: crystal structure of quee from burkholderia multivorans in complex2 with adomet, 7-carboxy-7-deazaguanine, and mg2+
|
|
|
| 58 | d1t9ba1 |
|
not modelled |
37.0 |
18 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
|
|
| 59 | c5ldgA_ |
|
not modelled |
36.4 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:(-)-isopiperitenone reductase;
PDBTitle: isopiperitenone reductase from mentha piperita in complex with2 isopiperitenone and nadp
|
|
|
| 60 | c2qasA_ |
|
not modelled |
36.1 |
20 |
PDB header:hydrolase activator
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of caulobacter crescentus sspb ortholog
|
|
|
| 61 | d2q4qa1 |
|
not modelled |
36.0 |
20 |
Fold:MTH938-like
Superfamily:MTH938-like
Family:MTH938-like |
|
|
| 62 | d1q6za1 |
|
not modelled |
35.7 |
19 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
|
|
| 63 | c6i7tB_ |
|
not modelled |
35.1 |
16 |
PDB header:translation
Chain: B: PDB Molecule:translation initiation factor eif-2b subunit alpha;
PDBTitle: eif2b:eif2 complex
|
|
|
| 64 | d1iuka_ |
|
not modelled |
34.9 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
| 65 | c5o30A_ |
|
not modelled |
34.6 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of the novel halohydrin dehalogenase hheg
|
|
|
| 66 | c3zlbA_ |
|
not modelled |
34.3 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoglycerate kinase;
PDBTitle: crystal structure of phosphoglycerate kinase from streptococcus2 pneumoniae
|
|
|
| 67 | c2a5hC_ |
|
not modelled |
33.6 |
17 |
PDB header:isomerase
Chain: C: PDB Molecule:l-lysine 2,3-aminomutase;
PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
|
|
|
| 68 | c5o3zK_ |
|
not modelled |
33.0 |
29 |
PDB header:oxidoreductase
Chain: K: PDB Molecule:sorbitol-6-phosphate dehydrogenase;
PDBTitle: crystal structure of sorbitol-6-phosphate 2-dehydrogenase srld from2 erwinia amylovora
|
|
|
| 69 | c3lqkA_ |
|
not modelled |
32.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit b;
PDBTitle: crystal structure of dipicolinate synthase subunit b from bacillus2 halodurans c
|
|
|
| 70 | c3qanB_ |
|
not modelled |
32.6 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:1-pyrroline-5-carboxylate dehydrogenase 1;
PDBTitle: crystal structure of 1-pyrroline-5-carboxylate dehydrogenase from2 bacillus halodurans
|
|
|
| 71 | c2qazC_ |
|
not modelled |
32.5 |
20 |
PDB header:hydrolase activator
Chain: C: PDB Molecule:sspb protein;
PDBTitle: structure of c. crescentus sspb ortholog
|
|
|
| 72 | c2qcnA_ |
|
not modelled |
31.6 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:uridine 5'-monophosphate synthase;
PDBTitle: covalent complex of the orotidine-5'-monophosphate decarboxylase2 domain of human ump synthase with 6-iodo-ump
|
|
|
| 73 | c3l7oB_ |
|
not modelled |
30.6 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from streptococcus2 mutans ua159
|
|
|
| 74 | d1yl7a1 |
|
not modelled |
30.6 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 75 | c5jg7A_ |
|
not modelled |
30.4 |
15 |
PDB header:metal transport
Chain: A: PDB Molecule:fur regulated salmonella iron transporter;
PDBTitle: crystal structure of putative periplasmic binding protein from2 salmonella typhimurium lt2
|
|
|
| 76 | c1w59B_ |
|
not modelled |
30.3 |
14 |
PDB header:cell division
Chain: B: PDB Molecule:cell division protein ftsz homolog 1;
PDBTitle: ftsz dimer, empty (m. jannaschii)
|
|
|
| 77 | c6gq0A_ |
|
not modelled |
30.2 |
11 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:putative sugar binding protein;
PDBTitle: crystal structure of ganp, a glucose-galactose binding protein from2 geobacillus stearothermophilus
|
|
|
| 78 | c5yaaD_ |
|
not modelled |
30.1 |
11 |
PDB header:hydrolase
Chain: D: PDB Molecule:meiosis regulator and mrna stability factor 1;
PDBTitle: crystal structure of marf1 nyn domain from mus musculus
|
|
|
| 79 | c4x54A_ |
|
not modelled |
29.9 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, short chain dehydrogenase/reductase family;
PDBTitle: crystal structure of an oxidoreductase (short chain2 dehydrogenase/reductase) from brucella ovis
|
|
|
| 80 | c3npgD_ |
|
not modelled |
29.9 |
12 |
PDB header:unknown function
Chain: D: PDB Molecule:uncharacterized duf364 family protein;
PDBTitle: crystal structure of a protein with unknown function from duf3642 family (ph1506) from pyrococcus horikoshii at 2.70 a resolution
|
|
|
| 81 | d1jeyb2 |
|
not modelled |
29.4 |
6 |
Fold:vWA-like
Superfamily:vWA-like
Family:Ku80 subunit N-terminal domain |
|
|
| 82 | c3vh3A_ |
|
not modelled |
29.2 |
27 |
PDB header:metal binding protein/protein transport
Chain: A: PDB Molecule:ubiquitin-like modifier-activating enzyme atg7;
PDBTitle: crystal structure of atg7ctd-atg8 complex
|
|
|
| 83 | c5ijgB_ |
|
not modelled |
29.1 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:cys/met metabolism pyridoxal-phosphate-dependent enzyme;
PDBTitle: crystal structure of o-acetylhomoserine sulfhydrolase from brucella2 melitensis at 2.0 a resolution
|
|
|
| 84 | c5sxyA_ |
|
not modelled |
28.8 |
10 |
PDB header:chaperone
Chain: A: PDB Molecule:bifunctional coenzyme pqq synthesis protein c/d;
PDBTitle: the solution nmr structure for the pqqd truncation of methylobacterium2 extorquens pqqcd representing a functional and stand-alone3 ribosomally synthesized and post-translational modified (ripp)4 recognition element (rre)
|
|
|
| 85 | c2j7pA_ |
|
not modelled |
28.7 |
20 |
PDB header:signal recognition
Chain: A: PDB Molecule:signal recognition particle protein;
PDBTitle: gmppnp-stabilized ng domain complex of the srp gtpases ffh2 and ftsy
|
|
|
| 86 | d16pka_ |
|
not modelled |
28.7 |
20 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
|
|
| 87 | c4ak9A_ |
|
not modelled |
28.4 |
16 |
PDB header:protein transport
Chain: A: PDB Molecule:cpftsy;
PDBTitle: structure of chloroplast ftsy from physcomitrella patens
|
|
|
| 88 | c4jenB_ |
|
not modelled |
28.4 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:cmp n-glycosidase;
PDBTitle: structure of clostridium botulinum cmp n-glycosidase, bcmb
|
|
|
| 89 | c6m9uA_ |
|
not modelled |
28.3 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, short chain dehydrogenase/reductase family
PDBTitle: structure of the apo-form of 20beta-hydroxysteroid dehydrogenase from2 bifidobacterium adolescentis strain l2-32
|
|
|
| 90 | d2djia1 |
|
not modelled |
27.8 |
16 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
|
|
| 91 | c4ldrA_ |
|
not modelled |
27.7 |
18 |
PDB header:isomerase, cell invasion
Chain: A: PDB Molecule:methylthioribose-1-phosphate isomerase;
PDBTitle: structure of the s283y mutant of mrdi
|
|
|
| 92 | c6qg0G_ |
|
not modelled |
27.6 |
21 |
PDB header:translation
Chain: G: PDB Molecule:translation initiation factor eif-2b subunit delta;
PDBTitle: structure of eif2b-eif2 (phosphorylated at ser51) complex (model 1)
|
|
|
| 93 | c4wt7B_ |
|
not modelled |
27.4 |
11 |
PDB header:transport protein
Chain: B: PDB Molecule:abc transporter substrate binding protein (ribose);
PDBTitle: crystal structure of an abc transporter solute binding protein2 (ipr025997) from agrobacterium vitis (avi_5165, target efi-511223)3 with bound allitol
|
|
|
| 94 | c3gx1A_ |
|
not modelled |
27.3 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin1832 protein;
PDBTitle: crystal structure of a domain of lin1832 from listeria innocua
|
|
|
| 95 | d2h1qa1 |
|
not modelled |
27.2 |
19 |
Fold:PLP-dependent transferase-like
Superfamily:Dhaf3308-like
Family:Dhaf3308-like |
|
|
| 96 | c3ecsD_ |
|
not modelled |
27.1 |
21 |
PDB header:translation
Chain: D: PDB Molecule:translation initiation factor eif-2b subunit alpha;
PDBTitle: crystal structure of human eif2b alpha
|
|
|
| 97 | c3hl2D_ |
|
not modelled |
26.9 |
15 |
PDB header:transferase
Chain: D: PDB Molecule:o-phosphoseryl-trna(sec) selenium transferase;
PDBTitle: the crystal structure of the human sepsecs-trnasec complex
|
|
|
| 98 | d1xw8a_ |
|
not modelled |
26.8 |
19 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:LamB/YcsF-like |
|
|
| 99 | c4zeoH_ |
|
not modelled |
26.8 |
27 |
PDB header:translation
Chain: H: PDB Molecule:translation initiation factor eif-2b-like protein;
PDBTitle: crystal structure of eif2b delta from chaetomium thermophilum
|
|
|
| 100 | c5hqjA_ |
|
not modelled |
26.7 |
12 |
PDB header:solute-binding protein
Chain: A: PDB Molecule:periplasmic binding protein/laci transcriptional regulator;
PDBTitle: crystal structure of abc transporter solute binding protein b1g1h72 from burkholderia graminis c4d1m, target efi-511179, in complex with3 d-arabinose
|
|
|
| 101 | c4gxtA_ |
|
not modelled |
26.7 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:a conserved functionally unknown protein;
PDBTitle: the crystal structure of a conserved functionally unknown protein from2 anaerococcus prevotii dsm 20548
|
|
|
| 102 | c3o26A_ |
|
not modelled |
26.7 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:salutaridine reductase;
PDBTitle: the structure of salutaridine reductase from papaver somniferum.
|
|
|
| 103 | d1jeya2 |
|
not modelled |
26.3 |
6 |
Fold:vWA-like
Superfamily:vWA-like
Family:Ku70 subunit N-terminal domain |
|
|
| 104 | c5ux5C_ |
|
not modelled |
26.2 |
16 |
PDB header:oxidoreductase/transferase
Chain: C: PDB Molecule:bifunctional protein proline utilization a (puta);
PDBTitle: structure of proline utilization a (puta) from corynebacterium2 freiburgense
|
|
|
| 105 | c2q5cA_ |
|
not modelled |
26.1 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:ntrc family transcriptional regulator;
PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
|
|
|
| 106 | d2gk3a1 |
|
not modelled |
26.0 |
10 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:STM3548-like |
|
|
| 107 | d2dlda2 |
|
not modelled |
25.5 |
24 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
|
|
| 108 | c4ru1C_ |
|
not modelled |
25.4 |
13 |
PDB header:transport protein
Chain: C: PDB Molecule:monosaccharide abc transporter substrate-binding protein,
PDBTitle: crystal structure of carbohydrate transporter acei_1806 from2 acidothermus cellulolyticus 11b, target efi-510965, in complex with3 myo-inositol
|
|
|
| 109 | c3upyB_ |
|
not modelled |
25.4 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of the brucella abortus enzyme catalyzing the first2 committed step of the methylerythritol 4-phosphate pathway.
|
|
|
| 110 | d1f0ka_ |
|
not modelled |
25.3 |
24 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Peptidoglycan biosynthesis glycosyltransferase MurG |
|
|
| 111 | d2dfaa1 |
|
not modelled |
25.2 |
27 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:LamB/YcsF-like |
|
|
| 112 | c4wzzA_ |
|
not modelled |
25.2 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:putative sugar abc transporter, substrate-binding protein;
PDBTitle: crystal structure of an abc transporter solute binding protein2 (ipr025997) from clostridium phytofermentas (cphy_0583, target efi-3 511148) with bound l-rhamnose
|
|
|
| 113 | c2nysA_ |
|
not modelled |
25.1 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:agr_c_3712p;
PDBTitle: x-ray crystal structure of protein agr_c_3712 from agrobacterium2 tumefaciens. northeast structural genomics consortium target atr88.
|
|
|
| 114 | d2nysa1 |
|
not modelled |
25.1 |
18 |
Fold:SspB-like
Superfamily:SspB-like
Family:AGR C 3712p-like |
|
|
| 115 | c2vpiA_ |
|
not modelled |
25.1 |
23 |
PDB header:ligase
Chain: A: PDB Molecule:gmp synthase;
PDBTitle: human gmp synthetase - glutaminase domain
|
|
|
| 116 | d2ozlb2 |
|
not modelled |
25.0 |
25 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain |
|
|
| 117 | d1ptma_ |
|
not modelled |
24.7 |
25 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PdxA-like |
|
|
| 118 | c4f9iA_ |
|
not modelled |
24.7 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:proline dehydrogenase/delta-1-pyrroline-5-carboxylate
PDBTitle: crystal structure of proline utilization a (puta) from geobacter2 sulfurreducens pca
|
|
|
| 119 | c3bmrA_ |
|
not modelled |
24.4 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pteridine reductase;
PDBTitle: structure of pteridine reductase 1 (ptr1) from trypanosoma2 brucei in ternary complex with cofactor (nadp+) and3 inhibitor (compound ax6)
|
|
|
| 120 | c5l53A_ |
|
not modelled |
24.1 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:(-)-menthone:(+)-neomenthol reductase;
PDBTitle: menthone neomenthol reductase from mentha piperita in complex with2 nadp
|
|
|