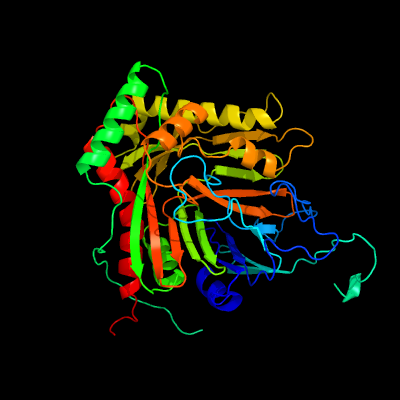
| 1 | c4penA_
|
|
 |
100.0 |
27 |
PDB header:lyase
Chain: A: PDB Molecule:anthranilate synthase component 1;
PDBTitle: structure of anthranilate synthase component i (trpe) from2 mycobacterium tuberculosis with inhibitor bound
|
|
|
|
| 2 | d1i7qa_
|
|
 |
100.0 |
24 |
Fold:ADC synthase
Superfamily:ADC synthase
Family:ADC synthase |
|
|
|
| 3 | d1i1qa_
|
|
 |
100.0 |
25 |
Fold:ADC synthase
Superfamily:ADC synthase
Family:ADC synthase |
|
|
|
| 4 | d1qdla_
|
|
 |
100.0 |
28 |
Fold:ADC synthase
Superfamily:ADC synthase
Family:ADC synthase |
|
|
|
| 5 | c5kckA_
|
|
 |
100.0 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:anthranilate synthase component i;
PDBTitle: crystal structure of anthranilate synthase component i from2 streptococcus pneumoniae tigr4
|
|
|
|
| 6 | c4grhA_
|
|
 |
100.0 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:aminodeoxychorismate synthase;
PDBTitle: crystal structure of pabb of stenotrophomonas maltophilia
|
|
|
|
| 7 | d2g5fa1
|
|
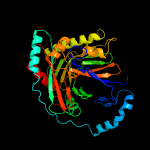 |
100.0 |
22 |
Fold:ADC synthase
Superfamily:ADC synthase
Family:ADC synthase |
|
|
|
| 8 | d1k0ga_
|
|
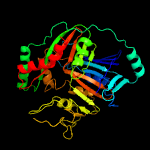 |
100.0 |
29 |
Fold:ADC synthase
Superfamily:ADC synthase
Family:ADC synthase |
|
|
|
| 9 | c2i6yA_
|
|
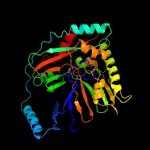 |
100.0 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:anthranilate synthase component i, putative;
PDBTitle: structure and mechanism of mycobacterium tuberculosis salicylate2 synthase, mbti
|
|
|
|
| 10 | d2fn0a1
|
|
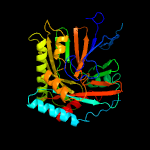 |
100.0 |
24 |
Fold:ADC synthase
Superfamily:ADC synthase
Family:ADC synthase |
|
|
|
| 11 | c3h9mA_
|
|
 |
100.0 |
27 |
PDB header:lyase
Chain: A: PDB Molecule:p-aminobenzoate synthetase, component i;
PDBTitle: crystal structure of para-aminobenzoate synthetase, component i from2 cytophaga hutchinsonii
|
|
|
|
| 12 | c3os6A_
|
|
 |
100.0 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:isochorismate synthase dhbc;
PDBTitle: crystal structure of putative 2,3-dihydroxybenzoate-specific2 isochorismate synthase, dhbc from bacillus anthracis.
|
|
|
|
| 13 | c3r74B_
|
|
 |
100.0 |
22 |
PDB header:lyase, biosynthetic protein
Chain: B: PDB Molecule:anthranilate/para-aminobenzoate synthases component i;
PDBTitle: crystal structure of 2-amino-2-desoxyisochorismate synthase (adic)2 synthase phze from burkholderia lata 383
|
|
|
|
| 14 | c3hwoB_
|
|
 |
100.0 |
20 |
PDB header:isomerase
Chain: B: PDB Molecule:isochorismate synthase entc;
PDBTitle: crystal structure of escherichia coli enterobactin-specific2 isochorismate synthase entc in complex with isochorismate
|
|
|
|
| 15 | d3bzna1
|
|
 |
100.0 |
23 |
Fold:ADC synthase
Superfamily:ADC synthase
Family:ADC synthase |
|
|
|
| 16 | c3gseA_
|
|
 |
100.0 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:menaquinone-specific isochorismate synthase;
PDBTitle: crystal structure of menaquinone-specific isochorismate synthase from2 yersinia pestis co92
|
|
|
|
| 17 | c3nqkA_
|
|
 |
26.5 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a structural genomics, unknown function2 (bacova_03322) from bacteroides ovatus at 2.61 a resolution
|
|
|
|
| 18 | d1cvra1
|
|
 |
24.0 |
19 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:Gingipain R (RgpB), C-terminal domain |
|
|
|
| 19 | d1rp3b_
|
|
 |
19.3 |
30 |
Fold:Non-globular all-alpha subunits of globular proteins
Superfamily:Anti-sigma factor FlgM
Family:Anti-sigma factor FlgM |
|
|
|
| 20 | c4k6nA_
|
|
 |
19.1 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:aminodeoxychorismate lyase;
PDBTitle: crystal structure of yeast 4-amino-4-deoxychorismate lyase
|
|
|
|
| 21 | c5jnoB_ |
|
not modelled |
16.1 |
24 |
PDB header:cell cycle
Chain: B: PDB Molecule:dna excision repair protein ercc-6-like;
PDBTitle: crystal structure of the bd1-ntpr complex from bend3 and pich
|
|
|
| 22 | c2km1A_ |
|
not modelled |
14.0 |
22 |
PDB header:protein binding
Chain: A: PDB Molecule:protein dre2;
PDBTitle: solution structure of the n-terminal domain of the yeast protein dre2
|
|
|
| 23 | c3vf0A_ |
|
not modelled |
13.2 |
17 |
PDB header:cell adhesion/protein binding
Chain: A: PDB Molecule:vinculin;
PDBTitle: raver1 in complex with metavinculin l954 deletion mutant
|
|
|
| 24 | d1hxra_ |
|
not modelled |
12.6 |
22 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:RabGEF Mss4 |
|
|
| 25 | d2fu5a1 |
|
not modelled |
12.0 |
22 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:RabGEF Mss4 |
|
|
| 26 | d1qkra_ |
|
not modelled |
11.6 |
17 |
Fold:Four-helical up-and-down bundle
Superfamily:alpha-catenin/vinculin-like
Family:alpha-catenin/vinculin |
|
|
| 27 | c4w5xA_ |
|
not modelled |
11.2 |
18 |
PDB header:viral protein
Chain: A: PDB Molecule:late protein h7;
PDBTitle: the structure of vaccina virus h7 protein displays a novel2 phosphoinositide binding fold required for membrane biogenesis
|
|
|
| 28 | c4aq2I_ |
|
not modelled |
10.0 |
27 |
PDB header:oxidoreductase
Chain: I: PDB Molecule:homogentisate 1,2-dioxygenase;
PDBTitle: resting state of homogentisate 1,2-dioxygenase
|
|
|
| 29 | c4oltA_ |
|
not modelled |
9.4 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:chitosanase;
PDBTitle: chitosanase complex structure
|
|
|
| 30 | c2hlwA_ |
|
not modelled |
8.9 |
17 |
PDB header:ligase, signaling protein
Chain: A: PDB Molecule:ubiquitin-conjugating enzyme e2 variant 1;
PDBTitle: solution structure of the human ubiquitin-conjugating2 enzyme variant uev1a
|
|
|
| 31 | d1xe7a_ |
|
not modelled |
8.7 |
41 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:YML079-like |
|
|
| 32 | c5f5tD_ |
|
not modelled |
8.2 |
62 |
PDB header:splicing
Chain: D: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the prp38-mfap1 complex of chaetomium2 thermophilum
|
|
|
| 33 | d2obpa1 |
|
not modelled |
8.2 |
38 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:ReutB4095-like |
|
|
| 34 | c2xt6B_ |
|
not modelled |
8.1 |
29 |
PDB header:lyase
Chain: B: PDB Molecule:2-oxoglutarate decarboxylase;
PDBTitle: crystal structure of mycobacterium smegmatis alpha-ketoglutarate2 decarboxylase homodimer (orthorhombic form)
|
|
|
| 35 | c1wazA_ |
|
not modelled |
7.9 |
57 |
PDB header:transport protein
Chain: A: PDB Molecule:merf;
PDBTitle: nmr structure determination of the bacterial mercury transporter,2 merf, in micelles
|
|
|
| 36 | c5foiB_ |
|
not modelled |
7.8 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:mycinamicin viii c21 methyl hydroxylase;
PDBTitle: crystal structure of mycinamicin viii c21 methyl hydroxylase mycci2 from micromonospora griseorubida bound to mycinamicin viii
|
|
|
| 37 | c1wqkA_ |
|
not modelled |
7.8 |
38 |
PDB header:toxin
Chain: A: PDB Molecule:toxin apetx1;
PDBTitle: solution structure of apetx1, a specific peptide inhibitor2 of human ether-a-go-go-related gene potassium channels3 from the venom of the sea anemone anthopleura4 elegantissima: a new fold for an herg toxin
|
|
|
| 38 | c2edgA_ |
|
not modelled |
7.7 |
23 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:glycine cleavage system h protein;
PDBTitle: solution structure of the gcv_h domain from mouse glycine
|
|
|
| 39 | d2ffca1 |
|
not modelled |
7.5 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
|
|
| 40 | c3dzaB_ |
|
not modelled |
7.3 |
32 |
PDB header:unknown function
Chain: B: PDB Molecule:uncharacterized putative membrane protein;
PDBTitle: crystal structure of a putative membrane protein of unknown function2 (yfdx) from klebsiella pneumoniae subsp. at 1.65 a resolution
|
|
|
| 41 | d1d5ta2 |
|
not modelled |
7.1 |
21 |
Fold:FAD-linked reductases, C-terminal domain
Superfamily:FAD-linked reductases, C-terminal domain
Family:GDI-like |
|
|
| 42 | c2lt2A_ |
|
not modelled |
6.4 |
16 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: nmr structure of ba42 protein from the psychrophilic bacteria bizionia2 argentinensis sp. nov.
|
|
|
| 43 | c4nm0B_ |
|
not modelled |
6.4 |
30 |
PDB header:transferase/peptide
Chain: B: PDB Molecule:axin-1;
PDBTitle: crystal structure of peptide inhibitor-free gsk-3/axin complex
|
|
|
| 44 | c3c66B_ |
|
not modelled |
6.3 |
30 |
PDB header:transferase
Chain: B: PDB Molecule:poly(a) polymerase;
PDBTitle: yeast poly(a) polymerase in complex with fip1 residues 80-105
|
|
|
| 45 | c3oaiB_ |
|
not modelled |
6.2 |
10 |
PDB header:membrane protein, cell adhesion
Chain: B: PDB Molecule:maltose-binding periplasmic protein, myelin protein p0;
PDBTitle: crystal structure of the extra-cellular domain of human myelin protein2 zero
|
|
|
| 46 | c4nm7B_ |
|
not modelled |
6.1 |
35 |
PDB header:transferase/peptide
Chain: B: PDB Molecule:axin-1;
PDBTitle: crystal structure of gsk-3/axin complex bound to phosphorylated wnt2 receptor lrp6 e-motif
|
|
|
| 47 | c4nm3B_ |
|
not modelled |
6.1 |
35 |
PDB header:transferase/peptide
Chain: B: PDB Molecule:axin-1;
PDBTitle: crystal structure of gsk-3/axin complex bound to phosphorylated n-2 terminal auto-inhibitory ps9 peptide
|
|
|
| 48 | c4nu1B_ |
|
not modelled |
6.1 |
35 |
PDB header:transferase/peptide
Chain: B: PDB Molecule:axin-1;
PDBTitle: crystal structure of a transition state mimic of the gsk-3/axin2 complex bound to phosphorylated n-terminal auto-inhibitory ps93 peptide
|
|
|
| 49 | c4nm5B_ |
|
not modelled |
6.1 |
35 |
PDB header:transferase/peptide
Chain: B: PDB Molecule:axin-1;
PDBTitle: crystal structure of gsk-3/axin complex bound to phosphorylated wnt2 receptor lrp6 c-motif
|
|
|
| 50 | d1eyba_ |
|
not modelled |
6.0 |
28 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Homogentisate dioxygenase |
|
|
| 51 | c1ey2A_ |
|
not modelled |
6.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:homogentisate 1,2-dioxygenase;
PDBTitle: human homogentisate dioxygenase with fe(ii)
|
|
|
| 52 | c4anrA_ |
|
not modelled |
5.6 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:soluble lytic transglycosylase b;
PDBTitle: crystal structure of soluble lytic transglycosylase sltb12 from pseudomonas aeruginosa
|
|
|
| 53 | d1zofa1 |
|
not modelled |
5.6 |
11 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
|
|
| 54 | d1ofcx1 |
|
not modelled |
5.6 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Myb/SANT domain |
|
|
| 55 | c6a2uA_ |
|
not modelled |
5.5 |
24 |
PDB header:signaling protein/oxidoreductase
Chain: A: PDB Molecule:twin-arginine translocation pathway signal;
PDBTitle: crystal structure of gamma-alpha subunit complex from burkholderia2 cepacia fad glucose dehydrogenase
|
|
|
| 56 | c6bkbE_ |
|
not modelled |
5.3 |
25 |
PDB header:immune system
Chain: E: PDB Molecule:polyprotein;
PDBTitle: structure of hepatitis c virus envelope glycoprotein e2 core from2 genotype 6a bound to broadly neutralizing antibody ar3a
|
|
|
| 57 | c5n81B_ |
|
not modelled |
5.3 |
16 |
PDB header:ligase
Chain: B: PDB Molecule:tyrocidine synthase 1;
PDBTitle: crystal structure of an engineered tyca variant in complex with an o-2 propargyl-beta-tyr-amp analog
|
|
|
| 58 | c5h1nB_ |
|
not modelled |
5.3 |
11 |
PDB header:unknown function
Chain: B: PDB Molecule:upf0253 protein yaep;
PDBTitle: crystal structure of sf173 from shigella flexneri
|
|
|
| 59 | d1ji8a_ |
|
not modelled |
5.3 |
20 |
Fold:DsrC, the gamma subunit of dissimilatory sulfite reductase
Superfamily:DsrC, the gamma subunit of dissimilatory sulfite reductase
Family:DsrC, the gamma subunit of dissimilatory sulfite reductase |
|
|
| 60 | c2h3oA_ |
|
not modelled |
5.3 |
57 |
PDB header:membrane protein
Chain: A: PDB Molecule:merf;
PDBTitle: structure of merft, a membrane protein with two trans-2 membrane helices
|
|
|
| 61 | c6mejC_ |
|
not modelled |
5.2 |
27 |
PDB header:immune system
Chain: C: PDB Molecule:e2 glycoprotein;
PDBTitle: broadly neutralizing antibodies against hcv use a cdrh3 disulfide2 motif to recognize an e2 glycoprotein site that can be targeted for3 vaccine design
|
|
|
| 62 | c6cg8A_ |
|
not modelled |
5.2 |
25 |
PDB header:dna binding protein/dna
Chain: A: PDB Molecule:upf0335 protein b7z12_12435;
PDBTitle: structure of c. crescentus gapr-dna
|
|
|
| 63 | d2v4jc1 |
|
not modelled |
5.1 |
18 |
Fold:DsrC, the gamma subunit of dissimilatory sulfite reductase
Superfamily:DsrC, the gamma subunit of dissimilatory sulfite reductase
Family:DsrC, the gamma subunit of dissimilatory sulfite reductase |
|
|
| 64 | c6od1B_ |
|
not modelled |
5.1 |
16 |
PDB header:signaling protein
Chain: B: PDB Molecule:anti-adapter protein irad;
PDBTitle: irad-bound to rssb d58p variant
|
|
|