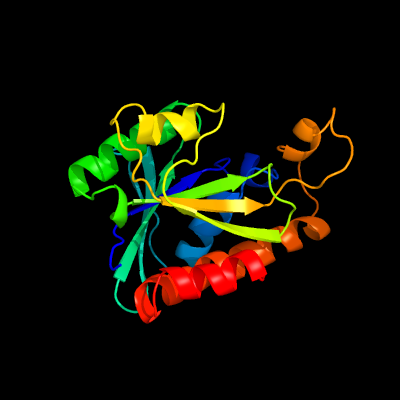
| 1 | c2mjlA_
|
|
 |
100.0 |
37 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: solution structure of peptidyl-trna hyrolase from vibrio cholerae
|
|
|
|
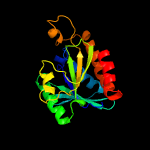
| 2 | c4dhwA_
|
|
 |
100.0 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: crystal structure of peptidyl-trna hydrolase from pseudomonas2 aeruginosa with adipic acid at 2.4 angstrom resolution
|
|
|
|
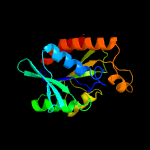
| 3 | d2ptha_
|
|
 |
100.0 |
40 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Peptidyl-tRNA hydrolase-like
Family:Peptidyl-tRNA hydrolase-like |
|
|
|
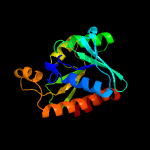
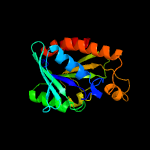
| 4 | c4fopA_
|
|
 |
100.0 |
39 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: crystal structure of peptidyl-trna hydrolase from acinetobacter2 baumannii at 1.86 a resolution
|
|
|
|
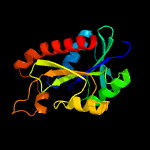
| 5 | c2z2jA_
|
|
 |
100.0 |
100 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: crystal structure of peptidyl-trna hydrolase from mycobacterium2 tuberculosis
|
|
|
|
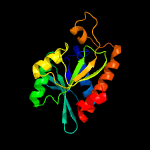
| 6 | c4q55B_
|
|
 |
100.0 |
31 |
PDB header:hydrolase
Chain: B: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: crystal structure of peptidyl-trna hydrolase from a gram-positive2 bacterium, streptococcus pyogenes at 2.19a resolution shows the3 closed structure of the substrate binding cleft
|
|
|
|
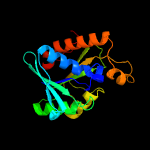
| 7 | c3neaA_
|
|
 |
100.0 |
38 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: crystal structure of peptidyl-trna hydrolase from francisella2 tularensis
|
|
|
|
| 8 | c3v2iA_
|
|
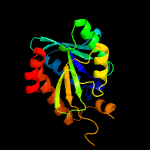 |
100.0 |
40 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: structure of a peptidyl-trna hydrolase (pth) from burkholderia2 thailandensis
|
|
|
|
| 9 | c4ylyB_
|
|
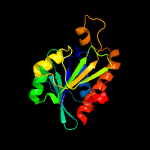 |
100.0 |
37 |
PDB header:hydrolase
Chain: B: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: crystal structure of peptidyl-trna hydrolase from a gram-positive2 bacterium, staphylococcus aureus at 2.25 angstrom resolution
|
|
|
|
| 10 | d1ryba_
|
|
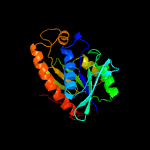 |
100.0 |
43 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Peptidyl-tRNA hydrolase-like
Family:Peptidyl-tRNA hydrolase-like |
|
|
|
| 11 | c5zx8A_
|
|
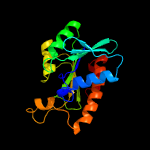 |
100.0 |
44 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: crystal structure of peptidyl-trna hydrolase from thermus thermophilus
|
|
|
|
| 12 | d1c8ba_
|
|
 |
81.6 |
25 |
Fold:Phosphorylase/hydrolase-like
Superfamily:HybD-like
Family:Germination protease |
|
|
|
| 13 | d1cfza_
|
|
 |
67.8 |
27 |
Fold:Phosphorylase/hydrolase-like
Superfamily:HybD-like
Family:Hydrogenase maturating endopeptidase HybD |
|
|
|
| 14 | c2e85B_
|
|
 |
67.7 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:hydrogenase 3 maturation protease;
PDBTitle: crystal structure of the hydrogenase 3 maturation protease
|
|
|
|
| 15 | c5zbyA_
|
|
 |
65.1 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:hydrogenase maturation protease hyci;
PDBTitle: crystal structure of a [nife] hydrogenase maturation protease hyci2 from thermococcus kodakarensis kod1
|
|
|
|
| 16 | c5ijaB_
|
|
 |
52.2 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:hydrogenase-specific maturation endopeptidase;
PDBTitle: [nife] hydrogenase maturation protease hybd from thermococcus2 kodakarensis
|
|
|
|
| 17 | c5ttxB_
|
|
 |
48.5 |
36 |
PDB header:hydrolase
Chain: B: PDB Molecule:hydrogenase 2 maturation peptidase;
PDBTitle: crystal structure of hydrogenase 2 maturation peptidase from2 thaumarchaeota archaeon scgc_ab-539-e09
|
|
|
|
| 18 | c2jvaA_
|
|
 |
41.1 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidyl-trna hydrolase domain protein;
PDBTitle: nmr solution structure of peptidyl-trna hydrolase domain protein from2 pseudomonas syringae pv. tomato. northeast structural genomics3 consortium target psr211
|
|
|
|
| 19 | d2ea9a1
|
|
 |
41.1 |
17 |
Fold:Profilin-like
Superfamily:YeeU-like
Family:YagB/YeeU/YfjZ-like |
|
|
|
| 20 | d2inwa1
|
|
 |
38.0 |
14 |
Fold:Profilin-like
Superfamily:YeeU-like
Family:YagB/YeeU/YfjZ-like |
|
|
|
| 21 | d2h28a1 |
|
not modelled |
36.6 |
17 |
Fold:Profilin-like
Superfamily:YeeU-like
Family:YagB/YeeU/YfjZ-like |
|
|
| 22 | c3tgnA_ |
|
not modelled |
34.9 |
17 |
PDB header:transcription
Chain: A: PDB Molecule:adc operon repressor adcr;
PDBTitle: crystal structure of the zinc-dependent marr family transcriptional2 regulator adcr in the zn(ii)-bound state
|
|
|
| 23 | c2jy9A_ |
|
not modelled |
33.5 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative trna hydrolase domain;
PDBTitle: nmr structure of putative trna hydrolase domain from2 salmonella typhimurium. northeast structural genomics3 consortium target str220
|
|
|
| 24 | c3mpoD_ |
|
not modelled |
28.7 |
15 |
PDB header:hydrolase
Chain: D: PDB Molecule:predicted hydrolase of the had superfamily;
PDBTitle: the crystal structure of a hydrolase from lactobacillus brevis
|
|
|
| 25 | c6ar6A_ |
|
not modelled |
28.2 |
25 |
PDB header:toxin/protein binding
Chain: A: PDB Molecule:toxin b;
PDBTitle: clostridioides difficile toxinb with dld-4 darpin
|
|
|
| 26 | c3ippA_ |
|
not modelled |
27.2 |
6 |
PDB header:transferase
Chain: A: PDB Molecule:putative thiosulfate sulfurtransferase ynje;
PDBTitle: crystal structure of sulfur-free ynje
|
|
|
| 27 | c3gygA_ |
|
not modelled |
26.4 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:ntd biosynthesis operon putative hydrolase ntdb;
PDBTitle: crystal structure of yhjk (haloacid dehalogenase-like hydrolase2 protein) from bacillus subtilis
|
|
|
| 28 | c6h98A_ |
|
not modelled |
23.9 |
6 |
PDB header:transferase
Chain: A: PDB Molecule:sulfurtransferase;
PDBTitle: native crystal structure of anaerobic ergothioneine biosynthesis2 enzyme from chlorobium limicola.
|
|
|
| 29 | c2bb3B_ |
|
not modelled |
23.7 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:cobalamin biosynthesis precorrin-6y methylase (cbie);
PDBTitle: crystal structure of cobalamin biosynthesis precorrin-6y methylase2 (cbie) from archaeoglobus fulgidus
|
|
|
| 30 | d2nqra3 |
|
not modelled |
20.9 |
18 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
|
|
| 31 | c1ujlA_ |
|
not modelled |
20.8 |
18 |
PDB header:membrane protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily h
PDBTitle: solution structure of the herg k+ channel s5-p2 extracellular linker
|
|
|
| 32 | c3ktdC_ |
|
not modelled |
20.5 |
27 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of a putative prephenate dehydrogenase (cgl0226)2 from corynebacterium glutamicum atcc 13032 at 2.60 a resolution
|
|
|
| 33 | c6fc1B_ |
|
not modelled |
19.9 |
32 |
PDB header:translation
Chain: B: PDB Molecule:protein eap1;
PDBTitle: crystal structure of the eif4e-eap1p complex from saccharomyces2 cerevisiae in the cap-bound state
|
|
|
| 34 | c5yhxH_ |
|
not modelled |
19.7 |
12 |
PDB header:metal binding protein
Chain: H: PDB Molecule:zinc transport transcriptional regulator;
PDBTitle: structure of lactococcus lactis zitr, wild type
|
|
|
| 35 | c1ynmA_ |
|
not modelled |
19.5 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:r.hinp1i restriction endonuclease;
PDBTitle: crystal structure of restriction endonuclease hinp1i
|
|
|
| 36 | c3hzuA_ |
|
not modelled |
19.5 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:thiosulfate sulfurtransferase ssea;
PDBTitle: crystal structure of probable thiosulfate sulfurtransferase ssea2 (rhodanese) from mycobacterium tuberculosis
|
|
|
| 37 | c3bj6B_ |
|
not modelled |
18.3 |
13 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: crystal structure of marr family transcription regulator sp03579
|
|
|
| 38 | d3eeqa2 |
|
not modelled |
17.6 |
20 |
Fold:CbiG N-terminal domain-like
Superfamily:CbiG N-terminal domain-like
Family:CbiG N-terminal domain-like |
|
|
| 39 | c4aq2I_ |
|
not modelled |
17.0 |
14 |
PDB header:oxidoreductase
Chain: I: PDB Molecule:homogentisate 1,2-dioxygenase;
PDBTitle: resting state of homogentisate 1,2-dioxygenase
|
|
|
| 40 | c5g2rA_ |
|
not modelled |
16.3 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:molybdopterin biosynthesis protein cnx1;
PDBTitle: crystal structure of the mo-insertase domain cnx1e from2 arabidopsis thaliana
|
|
|
| 41 | c3utnX_ |
|
not modelled |
16.2 |
19 |
PDB header:transferase
Chain: X: PDB Molecule:thiosulfate sulfurtransferase tum1;
PDBTitle: crystal structure of tum1 protein from saccharomyces cerevisiae
|
|
|
| 42 | c3kp3B_ |
|
not modelled |
16.0 |
14 |
PDB header:transcription regulator/antibiotic
Chain: B: PDB Molecule:transcriptional regulator tcar;
PDBTitle: staphylococcus epidermidis in complex with ampicillin
|
|
|
| 43 | d1am7a_ |
|
not modelled |
15.1 |
17 |
Fold:Lysozyme-like
Superfamily:Lysozyme-like
Family:Lambda lysozyme |
|
|
| 44 | d2bb3a1 |
|
not modelled |
14.8 |
29 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
| 45 | c3e8mD_ |
|
not modelled |
14.6 |
19 |
PDB header:transferase
Chain: D: PDB Molecule:acylneuraminate cytidylyltransferase;
PDBTitle: structure-function analysis of 2-keto-3-deoxy-d-glycero-d-galacto-2 nononate-9-phosphate (kdn) phosphatase defines a new clad within the3 type c0 had subfamily
|
|
|
| 46 | c5jlsA_ |
|
not modelled |
14.3 |
24 |
PDB header:transcription
Chain: A: PDB Molecule:adhesin competence repressor;
PDBTitle: crystal structure of adhesin competence repressor (adcr) from2 streptococcus pyogenes (c-terminally his tagged)
|
|
|
| 47 | c4uz2D_ |
|
not modelled |
14.0 |
19 |
PDB header:hydrolase
Chain: D: PDB Molecule:cell wall-binding endopeptidase-related protein;
PDBTitle: crystal structure of the n-terminal lysm domains from the putative2 nlpc/p60 d,l endopeptidase from t. thermophilus
|
|
|
| 48 | c3wz2C_ |
|
not modelled |
13.7 |
23 |
PDB header:chaperone
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of pyrococcus furiosus pbaa, an archaeal homolog of2 proteasome-assembly chaperone
|
|
|
| 49 | d1lama2 |
|
not modelled |
13.7 |
30 |
Fold:Macro domain-like
Superfamily:Macro domain-like
Family:Leucine aminopeptidase (Aminopeptidase A), N-terminal domain |
|
|
| 50 | c3n1uA_ |
|
not modelled |
13.5 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:hydrolase, had superfamily, subfamily iii a;
PDBTitle: structure of putative had superfamily (subfamily iii a) hydrolase from2 legionella pneumophila
|
|
|
| 51 | d2bgxa2 |
|
not modelled |
13.4 |
27 |
Fold:N-acetylmuramoyl-L-alanine amidase-like
Superfamily:N-acetylmuramoyl-L-alanine amidase-like
Family:N-acetylmuramoyl-L-alanine amidase-like |
|
|
| 52 | d1s2oa1 |
|
not modelled |
13.2 |
19 |
Fold:HAD-like
Superfamily:HAD-like
Family:Predicted hydrolases Cof |
|
|
| 53 | c3s2wB_ |
|
not modelled |
13.1 |
13 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: the crystal structure of a marr transcriptional regulator from2 methanosarcina mazei go1
|
|
|
| 54 | c3vdoB_ |
|
not modelled |
13.0 |
11 |
PDB header:dna binding protein/protein binding
Chain: B: PDB Molecule:anti-sigma-k factor rska;
PDBTitle: structure of extra-cytoplasmic function(ecf) sigma factor sigk in2 complex with its negative regulator rska from mycobacterium3 tuberculosis
|
|
|
| 55 | d1bofa2 |
|
not modelled |
12.7 |
38 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:G proteins |
|
|
| 56 | c3bpqD_ |
|
not modelled |
12.6 |
36 |
PDB header:toxin
Chain: D: PDB Molecule:toxin rele3;
PDBTitle: crystal structure of relb-rele antitoxin-toxin complex from2 methanococcus jannaschii
|
|
|
| 57 | c3av6A_ |
|
not modelled |
12.2 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:dna (cytosine-5)-methyltransferase 1;
PDBTitle: crystal structure of mouse dna methyltransferase 1 with adomet
|
|
|
| 58 | c1uarA_ |
|
not modelled |
11.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:rhodanese;
PDBTitle: crystal structure of rhodanese from thermus thermophilus hb8
|
|
|
| 59 | c3ecoB_ |
|
not modelled |
11.3 |
3 |
PDB header:transcription
Chain: B: PDB Molecule:mepr;
PDBTitle: crystal structure of mepr, a transcription regulator of the2 staphylococcus aureus multidrug efflux pump mepa
|
|
|
| 60 | c2jcyA_ |
|
not modelled |
11.3 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:1-deoxy-d-xylulose 5-phosphate reductoisomerase;
PDBTitle: x-ray structure of mutant 1-deoxy-d-xylulose 5-phosphate2 reductoisomerase, dxr, rv2870c, from mycobacterium tuberculosis
|
|
|
| 61 | c6alyA_ |
|
not modelled |
11.1 |
19 |
PDB header:transcription
Chain: A: PDB Molecule:mediator of rna polymerase ii transcription subunit 15;
PDBTitle: solution structure of yeast med15 abd2 residues 277-368
|
|
|
| 62 | d2al3a1 |
|
not modelled |
11.0 |
9 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:UBX domain |
|
|
| 63 | c4ol9A_ |
|
not modelled |
10.7 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative 2-dehydropantoate 2-reductase;
PDBTitle: crystal structure of putative 2-dehydropantoate 2-reductase pane from2 mycobacterium tuberculosis complexed with nadp and oxamate
|
|
|
| 64 | c2p9jH_ |
|
not modelled |
10.5 |
23 |
PDB header:structural genomics, unknown function
Chain: H: PDB Molecule:hypothetical protein aq2171;
PDBTitle: crystal structure of aq2171 from aquifex aeolicus
|
|
|
| 65 | c1uz5A_ |
|
not modelled |
10.3 |
23 |
PDB header:molybdopterin biosynthesis
Chain: A: PDB Molecule:402aa long hypothetical molybdopterin
PDBTitle: the crystal structure of molybdopterin biosynthesis moea2 protein from pyrococcus horikosii
|
|
|
| 66 | c5t57A_ |
|
not modelled |
10.3 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:semialdehyde dehydrogenase nad-binding protein;
PDBTitle: crystal structure of a semialdehyde dehydrogenase nad-binding protein2 from cupriavidus necator in complex with calcium and nad
|
|
|
| 67 | d2etha1 |
|
not modelled |
10.2 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
|
|
| 68 | c2bh7A_ |
|
not modelled |
9.9 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-acetylmuramoyl-l-alanine amidase;
PDBTitle: crystal structure of a semet derivative of amid at 2.22 angstroms
|
|
|
| 69 | d1k47a2 |
|
not modelled |
9.8 |
7 |
Fold:Ferredoxin-like
Superfamily:GHMP Kinase, C-terminal domain
Family:Phosphomevalonate kinase (PMK) |
|
|
| 70 | c6oq5A_ |
|
not modelled |
9.8 |
25 |
PDB header:toxin
Chain: A: PDB Molecule:toxin b;
PDBTitle: structure of the full-length clostridium difficile toxin b in complex2 with 3 vhhs
|
|
|
| 71 | c3mn1B_ |
|
not modelled |
9.7 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable yrbi family phosphatase;
PDBTitle: crystal structure of probable yrbi family phosphatase from pseudomonas2 syringae pv.phaseolica 1448a
|
|
|
| 72 | c4r04A_ |
|
not modelled |
9.4 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:toxin a;
PDBTitle: clostridium difficile toxin a (tcda)
|
|
|
| 73 | d1s3ja_ |
|
not modelled |
9.2 |
28 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
|
|
| 74 | c3fzqA_ |
|
not modelled |
9.1 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative hydrolase;
PDBTitle: crystal structure of putative haloacid dehalogenase-like hydrolase2 (yp_001086940.1) from clostridium difficile 630 at 2.10 a resolution
|
|
|
| 75 | d1k1ea_ |
|
not modelled |
9.0 |
23 |
Fold:HAD-like
Superfamily:HAD-like
Family:Probable phosphatase YrbI |
|
|
| 76 | c1okgA_ |
|
not modelled |
8.7 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:possible 3-mercaptopyruvate sulfurtransferase;
PDBTitle: 3-mercaptopyruvate sulfurtransferase from leishmania major
|
|
|
| 77 | c3rfqC_ |
|
not modelled |
8.6 |
14 |
PDB header:biosynthetic protein
Chain: C: PDB Molecule:pterin-4-alpha-carbinolamine dehydratase moab2;
PDBTitle: crystal structure of pterin-4-alpha-carbinolamine dehydratase moab22 from mycobacterium marinum
|
|
|
| 78 | c3k96B_ |
|
not modelled |
8.6 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glycerol-3-phosphate dehydrogenase [nad(p)+];
PDBTitle: 2.1 angstrom resolution crystal structure of glycerol-3-phosphate2 dehydrogenase (gpsa) from coxiella burnetii
|
|
|
| 79 | c4ztuA_ |
|
not modelled |
8.5 |
22 |
PDB header:dna binding protein/dna
Chain: A: PDB Molecule:dna polymerase subunit gamma-1;
PDBTitle: structural basis for processivity and antiviral drug toxicity in human2 mitochondrial dna replicase
|
|
|
| 80 | c4fhtA_ |
|
not modelled |
8.5 |
23 |
PDB header:transcription
Chain: A: PDB Molecule:pcav transcriptional regulator;
PDBTitle: crystal structure of the pcav transcriptional regulator from2 streptomyces coelicolor in complex with its natural ligand
|
|
|
| 81 | c4navB_ |
|
not modelled |
8.4 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:hypothetical protein xcc279;
PDBTitle: crystal structure of hypothetical protein xcc2798 from xanthomonas2 campestris, target efi-508608
|
|
|
| 82 | c4bolA_ |
|
not modelled |
8.3 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:ampdh2;
PDBTitle: crystal structure of ampdh2 from pseudomonas aeruginosa in2 complex with pentapeptide
|
|
|
| 83 | d3broa1 |
|
not modelled |
8.2 |
7 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
|
|
| 84 | c5cvoA_ |
|
not modelled |
8.1 |
23 |
PDB header:hydrolase/protein binding
Chain: A: PDB Molecule:wd repeat-containing protein 48;
PDBTitle: wdr48:usp46~ubiquitin ternary complex
|
|
|
| 85 | c4bndB_ |
|
not modelled |
8.1 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:alpha-phosphoglucomutase;
PDBTitle: structure of an atypical alpha-phosphoglucomutase similar to2 eukaryotic phosphomannomutases
|
|
|
| 86 | c6czlA_ |
|
not modelled |
8.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:atp phosphoribosyltransferase catalytic subunit;
PDBTitle: crystal structure of medicago truncatula atp-phosphoribosyltransferase2 in relaxed form
|
|
|
| 87 | d1fima_ |
|
not modelled |
7.9 |
17 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:MIF-related |
|
|
| 88 | c6jbxB_ |
|
not modelled |
7.8 |
13 |
PDB header:transcription/dna
Chain: B: PDB Molecule:fatty acid biosynthesis transcriptional regulator;
PDBTitle: crystal structure of streptococcus pneumoniae fabt in complex with dna
|
|
|
| 89 | c3oopA_ |
|
not modelled |
7.8 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin2960 protein;
PDBTitle: the structure of a protein with unknown function from listeria innocua2 clip11262
|
|
|
| 90 | d1kmma1 |
|
not modelled |
7.7 |
15 |
Fold:Anticodon-binding domain-like
Superfamily:Class II aaRS ABD-related
Family:Anticodon-binding domain of Class II aaRS |
|
|
| 91 | d1wdea_ |
|
not modelled |
7.6 |
40 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
| 92 | c3ikmD_ |
|
not modelled |
7.4 |
22 |
PDB header:transferase
Chain: D: PDB Molecule:dna polymerase subunit gamma-1;
PDBTitle: crystal structure of human mitochondrial dna polymerase holoenzyme
|
|
|
| 93 | c3boqB_ |
|
not modelled |
7.4 |
10 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, marr family;
PDBTitle: crystal structure of marr family transcriptional regulator from2 silicibacter pomeroyi
|
|
|
| 94 | c2r8zC_ |
|
not modelled |
7.4 |
27 |
PDB header:hydrolase
Chain: C: PDB Molecule:3-deoxy-d-manno-octulosonate 8-phosphate phosphatase;
PDBTitle: crystal structure of yrbi phosphatase from escherichia coli in complex2 with a phosphate and a calcium ion
|
|
|
| 95 | c3kbqA_ |
|
not modelled |
7.4 |
28 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein ta0487;
PDBTitle: the crystal structure of the protein cina with unknown function from2 thermoplasma acidophilum
|
|
|
| 96 | c3ihtB_ |
|
not modelled |
7.3 |
40 |
PDB header:transferase
Chain: B: PDB Molecule:s-adenosyl-l-methionine methyl transferase;
PDBTitle: crystal structure of s-adenosyl-l-methionine methyl transferase2 (yp_165822.1) from silicibacter pomeroyi dss-3 at 1.80 a resolution
|
|
|
| 97 | c2avxA_ |
|
not modelled |
7.3 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:regulatory protein sdia;
PDBTitle: solution structure of e coli sdia1-171
|
|
|
| 98 | c2abyA_ |
|
not modelled |
7.2 |
22 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein ta0743;
PDBTitle: solution structure of ta0743 from thermoplasma acidophilum
|
|
|
| 99 | c2cukC_ |
|
not modelled |
7.2 |
35 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glycerate dehydrogenase/glyoxylate reductase;
PDBTitle: crystal structure of tt0316 protein from thermus thermophilus hb8
|
|
|